Extracellular 5'-methylthioadenosine inhibits intracellular symmetric dimethylarginine protein methylation of FUSE-binding proteins
- PMID: 35963436
- PMCID: PMC9467882
- DOI: 10.1016/j.jbc.2022.102367
Extracellular 5'-methylthioadenosine inhibits intracellular symmetric dimethylarginine protein methylation of FUSE-binding proteins
Abstract
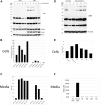
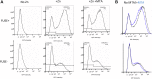
Methylthioadenosine phosphorylase (MTAP) is a key enzyme in the methionine salvage pathway that converts the polyamine synthesis byproduct 5'-deoxy-5'-methylthioadenosine (MTA) into methionine. Inactivation of MTAP, often by homozygous deletion, is found in both solid and hematologic malignancies and is one of the most frequently observed genetic alterations in human cancer. Previous work established that MTAP-deleted cells accumulate MTA and contain decreased amounts of proteins with symmetric dimethylarginine (sDMA). These findings led to the hypothesis that accumulation of intracellular MTA inhibits the protein arginine methylase (PRMT5) responsible for bulk protein sDMAylation. Here, we confirm that MTAP-deleted cells have increased MTA accumulation and reduced protein sDMAylation. However, we also show that addition of extracellular MTA can cause a dramatic reduction of the steady-state levels of sDMA-containing proteins in MTAP+ cells, even though no sustained increase in intracellular MTA is found because of catabolism of MTA by MTAP. We determined that inhibition of protein sDMAylation by MTA occurs within 48 h, is reversible, and is specific. In addition, we have identified two enhancer-binding proteins, FUBP1 and FUBP3, that are differentially sDMAylated in response to MTAP and MTA. These proteins work via the far upstream element site located upstream of Myc and other promoters. Using a transcription reporter construct containing the far upstream element site, we demonstrate that MTA addition can reduce transcription, suggesting that the reduction in FUBP1 and FUBP3 sDMAylation has functional consequences. Overall, our findings show that extracellular MTA can inhibit protein sDMAylation and that this inhibition can affect FUBP function.
Keywords: DNA-binding protein; metabolism; methionine; protein methylation; proteomics.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures





References
-
- Toohey J.I. Methylthioadenosine nucleoside phosphorylase deficiency in methylthio-dependent cancer cells. Biochem. Biophys. Res. Commun. 1978;83:27–35. - PubMed
-
- Cairns P., Polascik T.J., Eby Y., Tokino K., Califano J., Merlo A., et al. Frequency of homozygous deletion at p16/CDKN2 in primary human tumours. Nat. Genet. 1995;11:210–212. - PubMed
-
- Pei J., Kruger W.D., Testa J.R. High-resolution analysis of 9p loss in human cancer cells using single nucleotide polymorphism-based mapping arrays. Cancer Genet. Cytogenet. 2006;170:65–68. - PubMed
-
- Kamatani N., Carson D.A. Abnormal regulation of methylthioadenosine and polyamine metabolism in methylthioadenosine phosphorylase-deficient human leukemic cell lines. Cancer Res. 1980;40:4178–4182. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

