Human eukaryotic initiation factor 4E (eIF4E) and the nucleotide-bound state of eIF4A regulate eIF4F binding to RNA
- PMID: 35963437
- PMCID: PMC9483636
- DOI: 10.1016/j.jbc.2022.102368
Human eukaryotic initiation factor 4E (eIF4E) and the nucleotide-bound state of eIF4A regulate eIF4F binding to RNA
Abstract
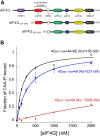
During translation initiation, the underlying mechanism by which the eukaryotic initiation factor (eIF) 4E, eIF4A, and eIF4G components of eIF4F coordinate their binding activities to regulate eIF4F binding to mRNA is poorly defined. Here, we used fluorescence anisotropy to generate thermodynamic and kinetic frameworks for the interaction of uncapped RNA with human eIF4F. We demonstrate that eIF4E binding to an autoinhibitory domain in eIF4G generates a high-affinity binding conformation of the eIF4F complex for RNA. In addition, we show that the nucleotide-bound state of the eIF4A component further regulates uncapped RNA binding by eIF4F, with a four-fold decrease in the equilibrium dissociation constant observed in the presence versus the absence of ATP. Monitoring uncapped RNA dissociation in real time reveals that ATP reduces the dissociation rate constant of RNA for eIF4F by ∼4-orders of magnitude. Thus, release of ATP from eIF4A places eIF4F in a dynamic state that has very fast association and dissociation rates from RNA. Monitoring the kinetic framework for eIF4A binding to eIF4G revealed two different rate constants that likely reflect two conformational states of the eIF4F complex. Furthermore, we determined that the eIF4G autoinhibitory domain promotes a more stable, less dynamic, eIF4A-binding state, which is overcome by eIF4E binding. Overall, our data support a model whereby eIF4E binding to eIF4G/4A stabilizes a high-affinity RNA-binding state of eIF4F and enables eIF4A to adopt a more dynamic interaction with eIF4G. This dynamic conformation may contribute to the ability of eIF4F to rapidly bind and release mRNA during scanning.
Keywords: RNA; cooperativity; eIF4A; eIF4E; eIF4F; eIF4G; translation initiation.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures




References
-
- Gingras A.C., Raught B., Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. - PubMed
-
- Abramson R.D., Dever T.E., Lawson T.G., Ray B.K., Thach R.E., Merrick W.C. The ATP-dependent interaction of eukaryotic initiation factors with mRNA. J. Biol. Chem. 1987;262:3826–3832. - PubMed
-
- Rogers G.W., Jr., Richter N.J., Lima W.F., Merrick W.C. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 2001;276:30914–30922. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

