Exploring the Energy Landscape of Riboswitches Using Collective Variables Based on Tertiary Contacts
- PMID: 35963460
- PMCID: PMC10042644
- DOI: 10.1016/j.jmb.2022.167788
Exploring the Energy Landscape of Riboswitches Using Collective Variables Based on Tertiary Contacts
Abstract
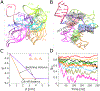
Messenger RNA regulatory elements, such as riboswitches, can display a high degree of flexibility. By characterizing their energy landscapes, and corresponding distributions of 3D configurations, structure-function relationships can be elucidated. Molecular dynamics simulation with enhanced sampling is an important strategy used to computationally access free energy landscapes characterizing the accessible 3D conformations of RNAs. While tertiary contacts are thought to play important roles in RNA dynamics, it is difficult, in explicit solvent, to sample the formation and breakage of tertiary contacts, such as helix-helix interactions, pseudoknot interactions, and junction interactions, while maintaining intact secondary structure elements. To this end, we extend previously developed collective variables and metadynamics efforts, to establish a simple metadynamics protocol, which utilizes only one collective variable, based on multiple tertiary contacts, to characterize the underlying free energy landscape of any RNA molecule. We develop a modified collective variable, the tertiary contacts distance (QTC), which can probe the formation and breakage of all or selectively chosen tertiary contacts of the RNA. The SAM-I riboswitch in the presence of three ionic and substrate conditions was investigated and validated against the structure ensemble previously generated using SAXS experiments. This efficient and easy to implement all-atom MD simulation based approach incorporating metadynamics to study RNA conformational dynamics can also be transferred to any other type of biomolecule.
Keywords: SAM-I Riboswitch; collective variable; contact distance; free energy; metadynamics.
Copyright © 2022. Published by Elsevier Ltd.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this pap
Figures
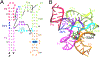

Similar articles
-
Hierarchical Conformational Dynamics Confers Thermal Adaptability to preQ1 RNA Riboswitches.J Mol Biol. 2020 Jul 24;432(16):4523-4543. doi: 10.1016/j.jmb.2020.06.002. Epub 2020 Jun 6. J Mol Biol. 2020. PMID: 32522558
-
The dynamic nature of RNA as key to understanding riboswitch mechanisms.Acc Chem Res. 2011 Dec 20;44(12):1339-48. doi: 10.1021/ar200035g. Epub 2011 Jun 16. Acc Chem Res. 2011. PMID: 21678902
-
Multiple conformations of SAM-II riboswitch detected with SAXS and NMR spectroscopy.Nucleic Acids Res. 2012 Apr;40(7):3117-30. doi: 10.1093/nar/gkr1154. Epub 2011 Dec 1. Nucleic Acids Res. 2012. PMID: 22139931 Free PMC article.
-
Structure-based insights into recognition and regulation of SAM-sensing riboswitches.Sci China Life Sci. 2023 Jan;66(1):31-50. doi: 10.1007/s11427-022-2188-7. Epub 2022 Nov 29. Sci China Life Sci. 2023. PMID: 36459353 Review.
-
Exploring RNA structure and dynamics through enhanced sampling simulations.Curr Opin Struct Biol. 2018 Apr;49:63-71. doi: 10.1016/j.sbi.2018.01.004. Epub 2018 Feb 4. Curr Opin Struct Biol. 2018. PMID: 29414513 Review.
Cited by
-
Accurate Characterization of Binding Kinetics and Allosteric Mechanisms for the HSP90 Chaperone Inhibitors Using AI-Augmented Integrative Biophysical Studies.JACS Au. 2024 Apr 1;4(4):1632-1645. doi: 10.1021/jacsau.4c00123. eCollection 2024 Apr 22. JACS Au. 2024. PMID: 38665669 Free PMC article.
-
Regulatory Mechanisms through RNA Conformational Switching and Dynamics.J Mol Biol. 2022 Sep 30;434(18):167794. doi: 10.1016/j.jmb.2022.167794. Epub 2022 Aug 18. J Mol Biol. 2022. PMID: 35988750 Free PMC article. No abstract available.
-
Embracing exascale computing in nucleic acid simulations.Curr Opin Struct Biol. 2024 Aug;87:102847. doi: 10.1016/j.sbi.2024.102847. Epub 2024 May 29. Curr Opin Struct Biol. 2024. PMID: 38815519 Free PMC article. Review.
-
Biophysical and Integrative Characterization of Protein Intrinsic Disorder as a Prime Target for Drug Discovery.Biomolecules. 2023 Mar 14;13(3):530. doi: 10.3390/biom13030530. Biomolecules. 2023. PMID: 36979465 Free PMC article. Review.
-
Cryo-EM reveals remodeling of a tandem riboswitch at 2.9 Å resolution.Res Sq [Preprint]. 2025 May 2:rs.3.rs-6422592. doi: 10.21203/rs.3.rs-6422592/v1. Res Sq. 2025. PMID: 40343338 Free PMC article. Preprint.
References
-
- Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG, Funnels, pathways, and the energy landscape of protein folding: a synthesis, Proteins 21 (3) (1995) 167–95. doi:10.1002/prot.340210302. URL http://www.ncbi.nlm.nih.gov/pubmed/7784423 - DOI - PubMed
-
- Clementi C, Nymeyer H, Onuchic JN, Topological and energetic factors: What determines the structural details of the transition state ensemble and “en-route” intermediates for protein folding? an investigation for small globular proteins, Journal of molecular biology 298 (5) (2000) 937–953. doi:10.1006/jmbi.2000.3693. - DOI - PubMed
-
- Whitford PC, Schug A, Saunders J, Hennelly SP, Onuchic JN, Sanbonmatsu KY, Nonlocal helix formation is key to understanding s-adenosylmethionine-1 riboswitch function, Biophys J 96 (2) (2009) L7–9. doi:10.1016/j.bpj.2008.10.033. URL https://www.ncbi.nlm.nih.gov/pubmed/19167285 - DOI - PMC - PubMed
-
- Whitford PC, Noel JK, Gosavi S, Schug A, Sanbonmatsu KY, Onuchic JN, An all-atom structure-based potential for proteins: bridging minimal models with all-atom empirical forcefields, Proteins 75 (2) (2009) 430–41. doi:10.1002/prot.22253. URL https://www.ncbi.nlm.nih.gov/pubmed/18837035 - DOI - PMC - PubMed
-
- Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, Donhofer A, Connell SR, Fucini P, Mielke T, Whitford PC, Onuchic JN, Yu Y, Sanbonmatsu KY, Hartmann RK, Penczek PA, Wilson DN, Spahn CM, Head swivel on the ribosome facilitates translocation by means of intra-subunit trna hybrid sites, Nature 468 (7324) (2010) 713–6. doi:10.1038/nature09547. URL https://www.ncbi.nlm.nih.gov/pubmed/21124459 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

