A method for Boolean analysis of protein interactions at a molecular level
- PMID: 35963857
- PMCID: PMC9375095
- DOI: 10.1038/s41467-022-32395-w
A method for Boolean analysis of protein interactions at a molecular level
Erratum in
-
Author Correction: A method for Boolean analysis of protein interactions at a molecular level.Nat Commun. 2023 Sep 6;14(1):5450. doi: 10.1038/s41467-023-41325-3. Nat Commun. 2023. PMID: 37673885 Free PMC article. No abstract available.
Abstract
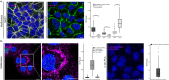
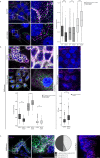
Determining the levels of protein-protein interactions is essential for the analysis of signaling within the cell, characterization of mutation effects, protein function and activation in health and disease, among others. Herein, we describe MolBoolean - a method to detect interactions between endogenous proteins in various subcellular compartments, utilizing antibody-DNA conjugates for identification and signal amplification. In contrast to proximity ligation assays, MolBoolean simultaneously indicates the relative abundances of protein A and B not interacting with each other, as well as the pool of A and B proteins that are proximal enough to be considered an AB complex. MolBoolean is applicable both in fixed cells and tissue sections. The specific and quantifiable data that the method generates provide opportunities for both diagnostic use and medical research.
© 2022. The Author(s).
Conflict of interest statement
The authors declare the following competing interests: O. Söderberg is the inventor of the MolBoolean method, with patent number: US2022042069A1-2022-02-10. The patent is now held by Atlas Antibodies, therefore, as employees of Atlas Antibodies, T.M. and C.K. declare financial interest. The remaining authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

