Genomic epidemiological analysis of Klebsiella pneumoniae from Portuguese hospitals reveals insights into circulating antimicrobial resistance
- PMID: 35963896
- PMCID: PMC9375070
- DOI: 10.1038/s41598-022-17996-1
Genomic epidemiological analysis of Klebsiella pneumoniae from Portuguese hospitals reveals insights into circulating antimicrobial resistance
Abstract
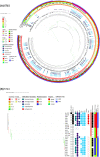
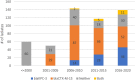
Klebsiella pneumoniae (Kp) bacteria are an increasing threat to public health and represent one of the most concerning pathogens involved in life-threatening infections and antimicrobial resistance (AMR). To understand the epidemiology of AMR of Kp in Portugal, we analysed whole genome sequencing, susceptibility testing and other meta data on 509 isolates collected nationwide from 16 hospitals and environmental settings between years 1980 and 2019. Predominant sequence types (STs) included ST15 (n = 161, 32%), ST147 (n = 36, 7%), ST14 (n = 26, 5%) or ST13 (n = 26, 5%), while 31% of isolates belonged to STs with fewer than 10 isolates. AMR testing revealed widespread resistance to aminoglycosides, fluoroquinolones, cephalosporins and carbapenems. The most common carbapenemase gene was blaKPC-3. Whilst the distribution of AMR linked plasmids appears uncorrelated with ST, their frequency has changed over time. Before year 2010, the dominant plasmid group was associated with the extended spectrum beta-lactamase gene blaCTX-M-15, but this group appears to have been displaced by another carrying the blaKPC-3 gene. Co-carriage of blaCTX-M and blaKPC-3 was uncommon. Our results from the largest genomics study of Kp in Portugal highlight the active transmission of strains with AMR genes and provide a baseline set of variants for future resistance monitoring and epidemiological studies.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in K. Pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. USA. 2015;112(27):E3574–E3581. doi: 10.1073/pnas.1501049112. - DOI - PMC - PubMed
-
- Redgrave, L.S., Sutton, S.B., Webber, M.A., & Piddock, L.J.V. Fluoroquinolone Resistance: Mechanisms, Impact on Bacteria, and Role in Evolutionary Success, Vol. 22, Trends in Microbiology 438–45 (Elsevier Ltd, 2014). 10.1016/j.tim.2014.04.007. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MR/M01360X/1/MRC_/Medical Research Council/United Kingdom
- BB/R013063/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/K000551/1/MRC_/Medical Research Council/United Kingdom
- MR/N010469/1/MRC_/Medical Research Council/United Kingdom
- MR/R020973/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources

