A composite strategy of genome-wide association study and copy number variation analysis for carcass traits in a Duroc pig population
- PMID: 35964005
- PMCID: PMC9375371
- DOI: 10.1186/s12864-022-08804-1
A composite strategy of genome-wide association study and copy number variation analysis for carcass traits in a Duroc pig population
Abstract
Background: Carcass traits are important in pig breeding programs for improving pork production. Understanding the genetic variants underlies complex phenotypes can help explain trait variation in pigs. In this study, we integrated a weighted single-step genome-wide association study (wssGWAS) and copy number variation (CNV) analyses to map genetic variations and genes associated with loin muscle area (LMA), loin muscle depth (LMD) and lean meat percentage (LMP) in Duroc pigs.
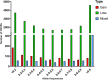
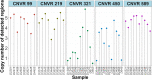
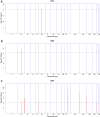
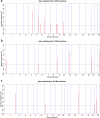
Results: Firstly, we performed a genome-wide analysis for CNV detection using GeneSeek Porcine SNP50 Bead chip data of 3770 pigs. A total of 11,100 CNVs were detected, which were aggregated by overlapping 695 CNV regions (CNVRs). Next, we investigated CNVs of pigs from the same population by whole-genome resequencing. A genome-wide analysis of 21 pigs revealed 23,856 CNVRs that were further divided into three categories (851 gain, 22,279 loss, and 726 mixed), which covered 190.8 Mb (~ 8.42%) of the pig autosomal genome. Further, the identified CNVRs were used to determine an overall validation rate of 68.5% for the CNV detection accuracy of chip data. CNVR association analyses identified one CNVR associated with LMA, one with LMD and eight with LMP after applying stringent Bonferroni correction. The wssGWAS identified eight, six and five regions explaining more than 1% of the additive genetic variance for LMA, LMD and LMP, respectively. The CNVR analyses and wssGWAS identified five common regions, of which three regions were associated with LMA and two with LMP. Four genes (DOK7, ARAP1, ELMO2 and SLC13A3) were highlighted as promising candidates according to their function.
Conclusions: We determined an overall validation rate for the CNV detection accuracy of low-density chip data and constructed a genomic CNV map for Duroc pigs using resequencing, thereby proving a value genetic variation resource for pig genome research. Furthermore, our study utilized a composite genetic strategy for complex traits in pigs, which will contribute to the study for elucidating the genetic architecture that may be influenced and regulated by multiple forms of variations.
Keywords: Carcass traits; Copy number variation; GWAS; Pigs.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Sanchez MP, Riquet J, Iannuccelli N, Gogué J, Billon Y, Demeure O, Caritez JC, Burgaud G, Fève K, Bonnet M, et al. Effects of quantitative trait loci on chromosomes 1, 2, 4, and 7 on growth, carcass, and meat quality traits in backcross Meishan x Large White pigs. J Anim Sci. 2006;84(3):526–537. doi: 10.2527/2006.843526x. - DOI - PubMed
MeSH terms
Grants and funding
- 2016YT03H062/Guangdong Yang Fan Innovative and Entrepreneurial Research Team Program
- 200-2018-XMZC-0001-107-0145/Guangdong Province Rural Revitalization Strategy Special Project
- 31972540/National Natural Science Foundation of China
- 2019BT02N630/Local Innovative and Research Teams Project of Guangdong Province
LinkOut - more resources
Full Text Sources
Miscellaneous

