Does swab type matter? Comparing methods for Mannheimia haemolytica recovery and upper respiratory microbiome characterization in feedlot cattle
- PMID: 35964128
- PMCID: PMC9375289
- DOI: 10.1186/s42523-022-00197-6
Does swab type matter? Comparing methods for Mannheimia haemolytica recovery and upper respiratory microbiome characterization in feedlot cattle
Abstract
Background: Bovine respiratory disease (BRD) is caused by interactions among host, environment, and pathogens. One standard method for antemortem pathogen identification in cattle with BRD is deep-guarded nasopharyngeal swabbing, which is challenging, costly, and waste generating. The objective was to compare the ability to recover Mannheimia haemolytica and compare microbial community structure using 29.5 inch (74.9 cm) deep-guarded nasopharyngeal swabs, 16 inch (40.6 cm) unguarded proctology swabs, or 6 inch (15.2 cm) unguarded nasal swabs when characterized using culture, real time-qPCR, and 16S rRNA gene sequencing. Samples for aerobic culture, qPCR, and 16S rRNA gene sequencing were collected from the upper respiratory tract of cattle 2 weeks after feedlot arrival.
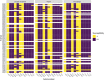
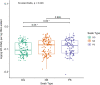
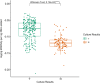
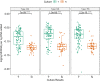
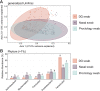
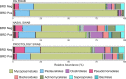
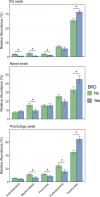
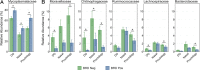
Results: There was high concordance of culture and qPCR results for all swab types (results for 77% and 81% of sampled animals completely across all 3 swab types for culture and qPCR respectively). Microbial communities were highly similar among samples collected with different swab types, and differences identified relative to treatment for BRD were also similar. Positive qPCR results for M. haemolytica were highly concordant (81% agreed completely), but samples collected by deep-guarded swabbing had lower amounts of Mh DNA identified (Kruskal-Wallis analysis of variance on ranks, P < 0.05; Dunn-test for pairwise comparison with Benjamini-Hochberg correction, P < 0.05) and lower frequency of positive compared to nasal and proctology swabs (McNemar's Chi-square test, P < 0.05).
Conclusions: Though differences existed among different types of swabs collected from individual cattle, nasal swabs and proctology swabs offer comparable results to deep-guarded nasopharyngeal swabs when identifying and characterizing M. haemolytica by culture, 16S rRNA gene sequencing, and qPCR.
Keywords: 16S rRNA gene sequencing; Antimicrobial resistance; Bovine respiratory disease; Culture; Disease surveillance; Metagenomics; qPCR.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- USDA-APHIS. U.S. feedlot processing practices for arriving cattle. National Animal Health Monitoring System, Feedlot 2011 Info Sheet. 2012.
LinkOut - more resources
Full Text Sources
