Polymyxins induce lipid scrambling and disrupt the homeostasis of Gram-negative bacteria membrane
- PMID: 35964158
- PMCID: PMC9515121
- DOI: 10.1016/j.bpj.2022.08.007
Polymyxins induce lipid scrambling and disrupt the homeostasis of Gram-negative bacteria membrane
Abstract
Polymyxins are increasingly used as the last-line therapeutic option for the treatment of infections caused by multidrug-resistant Gram-negative bacteria. However, efforts to address the resistance in superbugs are compromised by a poor understanding of the bactericidal modes because high-resolution detection of the cell structure is still lacking. By performing molecular dynamics simulations at a coarse-grained level, here we show that polymyxin B (PmB) disrupts Gram-negative bacterial membranes by altering lipid homeostasis and asymmetry. We found that the binding of PmBs onto the asymmetric outer membrane (OM) loosens the packing of lipopolysaccharides (LPS) and induces unbalanced bending torque between the inner and outer leaflets, which in turn triggers phospholipids to flip from the inner leaflet to the outer leaflet to compensate for the stress deformation. Meanwhile, some LPSs may be detained on the inner membrane (IM). Then, the lipid-scrambled OM undergoes phase separation. Defects are created at the boundaries between LPS-rich domains and phospholipid-rich domains, which consequently facilitate the uptake of PmB across the OM. Finally, PmBs target LPSs detained on the IM and similarly perturb the IM. This lipid Scramble, membrane phase Separation, and peptide Translocation model depicts a novel mechanism by which polymyxins kill bacteria and sheds light on developing a new generation of polymyxins or antibiotic adjuvants with improved killing activities and higher therapeutic indices.
Copyright © 2022 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
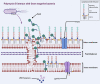



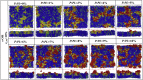
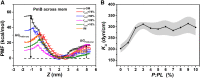

References
-
- Maria-Neto S., De Almeida K.C., et al. Franco O.L. Understanding bacterial resistance to antimicrobial peptides: from the surface to deep inside. Biochim. Biophys. Acta. 2015;1848:3078–3088. - PubMed
-
- Magana M., Pushpanathan M., et al. Tegos G.P. The value of antimicrobial peptides in the age of resistance. Lancet. Infect. Lancet Infect. Dis. 2020;20:e216–e230. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

