Comprehensive characterization of immune landscape of Indian and Western triple negative breast cancers
- PMID: 35964339
- PMCID: PMC9386467
- DOI: 10.1016/j.tranon.2022.101511
Comprehensive characterization of immune landscape of Indian and Western triple negative breast cancers
Erratum in
-
Corrigendum to "Comprehensive characterization of immune landscape of Indian and Western triple negative breast cancers": Translational Oncology 2022 Nov; 25:101511.Transl Oncol. 2023 Jan;27:101574. doi: 10.1016/j.tranon.2022.101574. Epub 2022 Nov 3. Transl Oncol. 2023. PMID: 36517200 Free PMC article. No abstract available.
Abstract
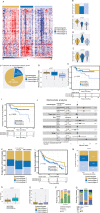
Purpose: Triple-negative breast cancer (TNBC) is a heterogeneous disease with a significant challenge to effectively manage in the clinic worldwide. Immunotherapy may be beneficial to TNBC patients if responders can be effectively identified. Here we sought to elucidate the immune landscape of TNBCs by stratifying patients into immune-specific subtypes (immunotypes) to decipher the molecular and cellular presentations and signaling events of this heterogeneous disease and associating them with their clinical outcomes and potential treatment options.
Experimental design: We profiled 730 immune genes in 88 retrospective Indian TNBC samples using the NanoString platform, established immunotypes using non-negative matrix factorization-based machine learning approach, and validated them using Western TNBCs (n=422; public datasets). Immunotype-specific gene signatures were associated with clinicopathological features, immune cell types, biological pathways, acute/chronic inflammatory responses, and immunogenic cell death processes. Responses to different immunotherapies associated with TNBC immunotypes were assessed using cross-cancer comparison to melanoma (n=504). Tumor-infiltrating lymphocytes (TILs) and pan-macrophage spatial marker expression were evaluated.
Results: We identified three robust transcriptome-based immunotypes in both Indian and Western TNBCs in similar proportions. Immunotype-1 tumors, mainly representing well-known claudin-low and immunomodulatory subgroups, harbored dense TIL infiltrates and T-helper-1 (Th1) response profiles associated with smaller tumors, pre-menopausal status, and a better prognosis. They displayed a cascade of events, including acute inflammation, damage-associated molecular patterns, T-cell receptor-related and chemokine-specific signaling, antigen presentation, and viral-mimicry pathways. On the other hand, immunotype-2 was enriched for Th2/Th17 responses, CD4+ regulatory cells, basal-like/mesenchymal immunotypes, and an intermediate prognosis. In contrast to the two T-cell enriched immunotypes, immunotype-3 patients expressed innate immune genes/proteins, including those representing myeloid infiltrations (validated by spatial immunohistochemistry), and had poor survival. Remarkably, a cross-cancer comparison analysis revealed the association of immunotype-1 with responses to anti-PD-L1 and MAGEA3 immunotherapies.
Conclusion: Overall, the TNBC immunotypes identified in TNBCs reveal different prognoses, immune infiltrations, signaling, acute/chronic inflammation leading to immunogenic cell death of cancer cells, and potentially distinct responses to immunotherapies. The overlap in immune characteristics in Indian and Western TNBCs suggests similar efficiency of immunotherapy in both populations if strategies to select patients according to immunotypes can be further optimized and implemented.
Keywords: Global oncology; Immune cells; Immune subtypes; Immunotherapy; India; Triple-negative breast cancer.
Copyright © 2022. Published by Elsevier Inc.
Conflict of interest statement
Declaration of Competing Interest AS has the following patents - Patient classification and prognostic method (GEP-NET) – Priority Patent – EP18425009.0, Patent – “Molecular predictors of therapeutic response to specific anti-cancer agents” (patent number US9506926B2). AS serves as a Scientific Advisor for Diagnostring Laboratories and Enedra Therapeutics and served as an Advisor for 4baseCare.
Figures






References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71(3):209–249. - PubMed
-
- Boyle P. Triple-negative breast cancer: epidemiological considerations and recommendations. Ann. Oncol. 2012;23(Suppl 6) vi7-12. - PubMed
-
- Bianchini G, De Angelis C, Licata L, Gianni L. Treatment landscape of triple-negative breast cancer - expanded options, evolving needs. Nat. Rev. Clin. Oncol. 2021 - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

