The diverse functions of FAT1 in cancer progression: good, bad, or ugly?
- PMID: 35965328
- PMCID: PMC9377080
- DOI: 10.1186/s13046-022-02461-8
The diverse functions of FAT1 in cancer progression: good, bad, or ugly?
Abstract
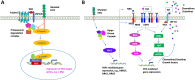
FAT atypical cadherin 1 (FAT1) is among the most frequently mutated genes in many types of cancer. Its highest mutation rate is found in head and neck squamous cell carcinoma (HNSCC), in which FAT1 is the second most frequently mutated gene. Thus, FAT1 has great potential to serve as a target or prognostic biomarker in cancer treatment. FAT1 encodes a member of the cadherin-like protein family. Under normal physiological conditions, FAT1 serves as a molecular "brake" on mitochondrial respiration and acts as a receptor for a signaling pathway regulating cell-cell contact interaction and planar cell polarity. In many cancers, loss of FAT1 function promotes epithelial-mesenchymal transition (EMT) and the formation of cancer initiation/stem-like cells. However, in some types of cancer, overexpression of FAT1 leads to EMT. The roles of FAT1 in cancer progression, which seems to be cancer-type specific, have not been clarified. To further study the function of FAT1 in cancers, this review summarizes recent relevant literature regarding this protein. In addition to phenotypic alterations due to FAT1 mutations, several signaling pathways and tumor immune systems known or proposed to be regulated by this protein are presented. The potential impact of detecting or targeting FAT1 mutations on cancer treatment is also prospectively discussed.
Keywords: Cancer progression; FAT1; Gene mutations; Signaling regulatory network; Targeted treatment.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Recent advances in the role of atypical cadherin FAT1 in tumorigenesis (Review).Oncol Lett. 2024 Dec 20;29(3):110. doi: 10.3892/ol.2024.14856. eCollection 2025 Mar. Oncol Lett. 2024. PMID: 39776648 Free PMC article. Review.
-
The Proteomic Landscape of Growth Factor Signaling Networks Associated with FAT1 Mutations in Head and Neck Cancers.Cancer Res. 2021 Sep 1;81(17):4402-4416. doi: 10.1158/0008-5472.CAN-20-3659. Epub 2021 Jun 24. Cancer Res. 2021. PMID: 34167951 Free PMC article.
-
Fat1 deletion promotes hybrid EMT state, tumour stemness and metastasis.Nature. 2021 Jan;589(7842):448-455. doi: 10.1038/s41586-020-03046-1. Epub 2020 Dec 16. Nature. 2021. PMID: 33328637 Free PMC article.
-
FAT1 knockdown enhances the CSC properties of HNSCC through p-CaMKII-mediated inactivation of the IFN pathway.Int J Biol Sci. 2025 Jan 1;21(2):671-684. doi: 10.7150/ijbs.95723. eCollection 2025. Int J Biol Sci. 2025. PMID: 39781458 Free PMC article.
-
Function and cancer genomics of FAT family genes (review).Int J Oncol. 2012 Dec;41(6):1913-8. doi: 10.3892/ijo.2012.1669. Epub 2012 Oct 17. Int J Oncol. 2012. PMID: 23076869 Free PMC article. Review.
Cited by
-
Exploration of prognostic biomarkers in head and neck squamous cell carcinoma microenvironment from TCGA database.Ann Transl Med. 2023 Feb 28;11(4):163. doi: 10.21037/atm-22-6481. Ann Transl Med. 2023. PMID: 36923087 Free PMC article.
-
Common skin cancers and their association with other non-cutaneous primary malignancies: a review of the literature.Med Oncol. 2024 May 17;41(6):157. doi: 10.1007/s12032-024-02385-7. Med Oncol. 2024. PMID: 38758457 Review.
-
Molecular Characterization of Atypical Fibroxanthoma and Pleomorphic Dermal Sarcoma.Cancers (Basel). 2025 May 27;17(11):1785. doi: 10.3390/cancers17111785. Cancers (Basel). 2025. PMID: 40507266 Free PMC article. Review.
-
Specific antigens in malignancy-associated membranous nephropathy.Front Med (Lausanne). 2024 Apr 15;11:1368457. doi: 10.3389/fmed.2024.1368457. eCollection 2024. Front Med (Lausanne). 2024. PMID: 38686366 Free PMC article. Review.
-
Recent advances in the role of atypical cadherin FAT1 in tumorigenesis (Review).Oncol Lett. 2024 Dec 20;29(3):110. doi: 10.3892/ol.2024.14856. eCollection 2025 Mar. Oncol Lett. 2024. PMID: 39776648 Free PMC article. Review.
References
-
- Campbell JD, Yau C, Bowlby R, Liu Y, Brennan K, Fan H, Taylor AM, Wang C, Walter V, Akbani R, Byers LA, Creighton CJ, Coarfa C, Shih J, Cherniack AD, Gevaert O, Prunello M, Shen H, Anur P, Chen J, Cheng H, Hayes DN, Bullman S, Pedamallu CS, Ojesina AI, Sadeghi S, Mungall KL, Robertson AG, Benz C, Schultz A, Kanchi RS, Gay CM, Hegde A, Diao L, Wang J, Ma W, Sumazin P, Chiu HS, Chen TW, Gunaratne P, Donehower L, Rader JS, Zuna R, Al-Ahmadie H, Lazar AJ, Flores ER, Tsai KY, Zhou JH, Rustgi AK, Drill E, Shen R, Wong CK, Cancer Genome Atlas Research N. Stuart JM, Laird PW, Hoadley KA, Weinstein JN, Peto M, Pickering CR, Chen Z, Van Waes C. Genomic, pathway network, and immunologic features distinguishing squamous carcinomas. Cell Rep. 2018;23(1):194–212. doi: 10.1016/j.celrep.2018.03.063. - DOI - PMC - PubMed
-
- Martin D, Degese MS, Vitale-Cross L, Iglesias-Bartolome R, Valera JLC, Wang Z, Feng X, Yeerna H, Vadmal V, Moroishi T, Thorne RF, Zaida M, Siegele B, Cheong SC, Molinolo AA, Samuels Y, Tamayo P, Guan KL, Lippman SM, Lyons JG, Gutkind JS. Assembly and activation of the Hippo signalome by FAT1 tumor suppressor. Nat Commun. 2018;9(1):2372. doi: 10.1038/s41467-018-04590-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

