CRISPR base editing of cis-regulatory elements enables the perturbation of neurodegeneration-linked genes
- PMID: 35965414
- PMCID: PMC9734028
- DOI: 10.1016/j.ymthe.2022.08.008
CRISPR base editing of cis-regulatory elements enables the perturbation of neurodegeneration-linked genes
Abstract
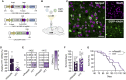
CRISPR technology has demonstrated broad utility for controlling target gene expression; however, there remains a need for strategies capable of modulating expression via the precise editing of non-coding regulatory elements. Here, we demonstrate that CRISPR base editors, a class of gene-modifying proteins capable of creating single-base substitutions in DNA, can be used to perturb gene expression via their targeted mutagenesis of cis-acting sequences. Using the promoter region of the human huntingtin (HTT) gene as an initial target, we show that editing of the binding site for the transcription factor NF-κB led to a marked reduction in HTT gene expression in base-edited cell populations. We found that these gene perturbations were persistent and specific, as a transcriptome-wide RNA analysis revealed minimal off-target effects resulting from the action of the base editor protein. We further demonstrate that this base-editing platform could influence gene expression in vivo as its delivery to a mouse model of Huntington's disease led to a potent decrease in HTT mRNA in striatal neurons. Finally, to illustrate the applicability of this concept, we target the amyloid precursor protein, showing that multiplex editing of its promoter region significantly perturbed its expression. These findings demonstrate the potential for base editors to regulate target gene expression.
Keywords: AAV; CRISPR; base editing; cis-regulatory elements; gene regulation.
Copyright © 2022 The American Society of Gene and Cell Therapy. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors have filed patent applications on CRISPR technologies.
Figures



Comment in
-
Tuning neurodegeneration-linked gene expression, one (edited) base at a time.Mol Ther. 2022 Dec 7;30(12):3512-3514. doi: 10.1016/j.ymthe.2022.10.003. Epub 2022 Oct 27. Mol Ther. 2022. PMID: 36302384 Free PMC article. No abstract available.
Similar articles
-
Strategies for Optimization of the Clustered Regularly Interspaced Short Palindromic Repeat-Based Genome Editing System for Enhanced Editing Specificity.Hum Gene Ther. 2022 Apr;33(7-8):358-370. doi: 10.1089/hum.2021.283. Epub 2022 Mar 1. Hum Gene Ther. 2022. PMID: 34963339 Review.
-
[Advances in base editing systems].Sheng Wu Gong Cheng Xue Bao. 2024 May 25;40(5):1271-1292. doi: 10.13345/j.cjb.230615. Sheng Wu Gong Cheng Xue Bao. 2024. PMID: 38783797 Review. Chinese.
-
Base Editing: The Ever Expanding Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) Tool Kit for Precise Genome Editing in Plants.Genes (Basel). 2020 Apr 24;11(4):466. doi: 10.3390/genes11040466. Genes (Basel). 2020. PMID: 32344599 Free PMC article. Review.
-
RNA-seq analysis reveals significant transcriptome changes in huntingtin-null human neuroblastoma cells.BMC Med Genomics. 2021 Jul 2;14(1):176. doi: 10.1186/s12920-021-01022-w. BMC Med Genomics. 2021. PMID: 34215255 Free PMC article.
-
Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors.Nature. 2019 May;569(7756):433-437. doi: 10.1038/s41586-019-1161-z. Epub 2019 Apr 17. Nature. 2019. PMID: 30995674 Free PMC article.
Cited by
-
Progress in and Prospects of Genome Editing Tools for Human Disease Model Development and Therapeutic Applications.Genes (Basel). 2023 Feb 14;14(2):483. doi: 10.3390/genes14020483. Genes (Basel). 2023. PMID: 36833410 Free PMC article. Review.
-
A high-fidelity CRISPR-Cas13 system improves abnormalities associated with C9ORF72-linked ALS/FTD.bioRxiv [Preprint]. 2023 Dec 12:2023.12.12.571328. doi: 10.1101/2023.12.12.571328. bioRxiv. 2023. Update in: Nat Commun. 2025 Jan 8;16(1):460. doi: 10.1038/s41467-024-55548-5. PMID: 38168370 Free PMC article. Updated. Preprint.
-
Targeting Duchenne muscular dystrophy by skipping DMD exon 45 with base editors.Mol Ther Nucleic Acids. 2023 Jul 27;33:572-586. doi: 10.1016/j.omtn.2023.07.029. eCollection 2023 Sep 12. Mol Ther Nucleic Acids. 2023. PMID: 37637209 Free PMC article.
-
Current trends in gene therapy to treat inherited disorders of the brain.Mol Ther. 2025 May 7;33(5):1988-2014. doi: 10.1016/j.ymthe.2025.03.057. Epub 2025 Apr 2. Mol Ther. 2025. PMID: 40181540 Review.
-
A high-fidelity CRISPR-Cas13 system improves abnormalities associated with C9ORF72-linked ALS/FTD.Nat Commun. 2025 Jan 8;16(1):460. doi: 10.1038/s41467-024-55548-5. Nat Commun. 2025. PMID: 39779681 Free PMC article.
References
-
- Buccitelli C., Selbach M. mRNAs, proteins and the emerging principles of gene expression control. Nat. Rev. Genet. 2020;21:630–644. - PubMed
-
- Wittkopp P.J., Kalay G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 2011;13:59–69. - PubMed
-
- Cramer P. Organization and regulation of gene transcription. Nature. 2019;573:45–54. - PubMed
-
- Yu Z., Pandian G.N., Hidaka T., Sugiyama H. Therapeutic gene regulation using pyrrole–imidazole polyamides. Adv. Drug Deliv. Rev. 2019;147:66–85. - PubMed
-
- Matharu N., Ahituv N. Modulating gene regulation to treat genetic disorders. Nat. Rev. Drug Discov. 2020;19:757–775. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

