CRISPR base editing of cis-regulatory elements enables the perturbation of neurodegeneration-linked genes
- PMID: 35965414
- PMCID: PMC9734028
- DOI: 10.1016/j.ymthe.2022.08.008
CRISPR base editing of cis-regulatory elements enables the perturbation of neurodegeneration-linked genes
Abstract
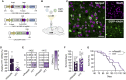
CRISPR technology has demonstrated broad utility for controlling target gene expression; however, there remains a need for strategies capable of modulating expression via the precise editing of non-coding regulatory elements. Here, we demonstrate that CRISPR base editors, a class of gene-modifying proteins capable of creating single-base substitutions in DNA, can be used to perturb gene expression via their targeted mutagenesis of cis-acting sequences. Using the promoter region of the human huntingtin (HTT) gene as an initial target, we show that editing of the binding site for the transcription factor NF-κB led to a marked reduction in HTT gene expression in base-edited cell populations. We found that these gene perturbations were persistent and specific, as a transcriptome-wide RNA analysis revealed minimal off-target effects resulting from the action of the base editor protein. We further demonstrate that this base-editing platform could influence gene expression in vivo as its delivery to a mouse model of Huntington's disease led to a potent decrease in HTT mRNA in striatal neurons. Finally, to illustrate the applicability of this concept, we target the amyloid precursor protein, showing that multiplex editing of its promoter region significantly perturbed its expression. These findings demonstrate the potential for base editors to regulate target gene expression.
Keywords: AAV; CRISPR; base editing; cis-regulatory elements; gene regulation.
Copyright © 2022 The American Society of Gene and Cell Therapy. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors have filed patent applications on CRISPR technologies.
Figures



Comment in
-
Tuning neurodegeneration-linked gene expression, one (edited) base at a time.Mol Ther. 2022 Dec 7;30(12):3512-3514. doi: 10.1016/j.ymthe.2022.10.003. Epub 2022 Oct 27. Mol Ther. 2022. PMID: 36302384 Free PMC article. No abstract available.
References
-
- Buccitelli C., Selbach M. mRNAs, proteins and the emerging principles of gene expression control. Nat. Rev. Genet. 2020;21:630–644. - PubMed
-
- Wittkopp P.J., Kalay G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 2011;13:59–69. - PubMed
-
- Cramer P. Organization and regulation of gene transcription. Nature. 2019;573:45–54. - PubMed
-
- Yu Z., Pandian G.N., Hidaka T., Sugiyama H. Therapeutic gene regulation using pyrrole–imidazole polyamides. Adv. Drug Deliv. Rev. 2019;147:66–85. - PubMed
-
- Matharu N., Ahituv N. Modulating gene regulation to treat genetic disorders. Nat. Rev. Drug Discov. 2020;19:757–775. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

