NAb-seq: an accurate, rapid, and cost-effective method for antibody long-read sequencing in hybridoma cell lines and single B cells
- PMID: 35965451
- PMCID: PMC9377246
- DOI: 10.1080/19420862.2022.2106621
NAb-seq: an accurate, rapid, and cost-effective method for antibody long-read sequencing in hybridoma cell lines and single B cells
Abstract
Despite their common use in research, monoclonal antibodies are currently not systematically sequenced. This can lead to issues with reproducibility and the occasional loss of antibodies with loss of cell lines. Hybridoma cell lines have been the primary means of generating monoclonal antibodies from immunized animals, including mice, rats, rabbits, and alpacas. Excluding therapeutic antibodies, few hybridoma-derived antibody sequences are known. Sanger sequencing has been "the gold standard" for antibody gene sequencing, but this method relies on the availability of species-specific degenerate primer sets for amplification of light and heavy antibody genes and it requires lengthy and expensive cDNA preparation. Here, we leveraged recent improvements in long-read Oxford Nanopore Technologies (ONT) sequencing to develop Nanopore Antibody sequencing (NAb-seq): a three-day, species-independent, and cost-effective workflow to characterize paired full-length immunoglobulin light- and heavy-chain genes from hybridoma cell lines. When compared to Sanger sequencing of two hybridoma cell lines, long-read ONT sequencing was highly accurate, reliable, and amenable to high throughput. We further show that the method is applicable to single cells, allowing efficient antibody discovery in rare populations such as memory B cells. In summary, NAb-seq promises to accelerate identification and validation of hybridoma antibodies as well as antibodies from single B cells used in research, diagnostics, and therapeutics.
Keywords: Antibody sequencing; B cell; hybridoma; long-read; nanopore sequencing; rat B cell cloning; single cell; workflow.
Conflict of interest statement
The authors report there are no competing interests to declare.
Figures
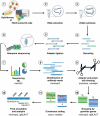
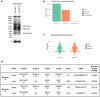


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
