Estimation of age-stratified contact rates during the COVID-19 pandemic using a novel inference algorithm
- PMID: 35965466
- PMCID: PMC9376725
- DOI: 10.1098/rsta.2021.0298
Estimation of age-stratified contact rates during the COVID-19 pandemic using a novel inference algorithm
Abstract
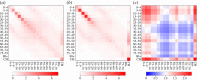
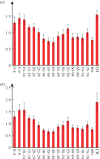
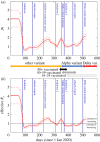
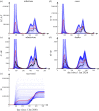
Well parameterized epidemiological models including accurate representation of contacts are fundamental to controlling epidemics. However, age-stratified contacts are typically estimated from pre-pandemic/peace-time surveys, even though interventions and public response likely alter contacts. Here, we fit age-stratified models, including re-estimation of relative contact rates between age classes, to public data describing the 2020-2021 COVID-19 outbreak in England. This data includes age-stratified population size, cases, deaths, hospital admissions and results from the Coronavirus Infection Survey (almost 9000 observations in all). Fitting stochastic compartmental models to such detailed data is extremely challenging, especially considering the large number of model parameters being estimated (over 150). An efficient new inference algorithm ABC-MBP combining existing approximate Bayesian computation (ABC) methodology with model-based proposals (MBPs) is applied. Modified contact rates are inferred alongside time-varying reproduction numbers that quantify changes in overall transmission due to pandemic response, and age-stratified proportions of asymptomatic cases, hospitalization rates and deaths. These inferences are robust to a range of assumptions including the values of parameters that cannot be estimated from available data. ABC-MBP is shown to enable reliable joint analysis of complex epidemiological data yielding consistent parametrization of dynamic transmission models that can inform data-driven public health policy and interventions. This article is part of the theme issue 'Technical challenges of modelling real-life epidemics and examples of overcoming these'.
Keywords: Bayesian inference; COVID-19; approximate Bayesian computation; contact matrix; model-based proposals; reproduction number.
Figures


Similar articles
-
Heterogeneity in the onwards transmission risk between local and imported cases affects practical estimates of the time-dependent reproduction number.Philos Trans A Math Phys Eng Sci. 2022 Oct 3;380(2233):20210308. doi: 10.1098/rsta.2021.0308. Epub 2022 Aug 15. Philos Trans A Math Phys Eng Sci. 2022. PMID: 35965464 Free PMC article.
-
Estimation of local time-varying reproduction numbers in noisy surveillance data.Philos Trans A Math Phys Eng Sci. 2022 Oct 3;380(2233):20210303. doi: 10.1098/rsta.2021.0303. Epub 2022 Aug 15. Philos Trans A Math Phys Eng Sci. 2022. PMID: 35965456 Free PMC article.
-
Refining epidemiological forecasts with simple scoring rules.Philos Trans A Math Phys Eng Sci. 2022 Oct 3;380(2233):20210305. doi: 10.1098/rsta.2021.0305. Epub 2022 Aug 15. Philos Trans A Math Phys Eng Sci. 2022. PMID: 35965461 Free PMC article.
-
Usage of Compartmental Models in Predicting COVID-19 Outbreaks.AAPS J. 2022 Sep 2;24(5):98. doi: 10.1208/s12248-022-00743-9. AAPS J. 2022. PMID: 36056223 Free PMC article. Review.
-
On the use of kernel approximate Bayesian computation to infer population history.Genes Genet Syst. 2015;90(3):153-62. doi: 10.1266/ggs.90.153. Genes Genet Syst. 2015. PMID: 26510570 Review.
Cited by
-
An algebraic framework for structured epidemic modelling.Philos Trans A Math Phys Eng Sci. 2022 Oct 3;380(2233):20210309. doi: 10.1098/rsta.2021.0309. Epub 2022 Aug 15. Philos Trans A Math Phys Eng Sci. 2022. PMID: 35965465 Free PMC article.
-
Visualization for epidemiological modelling: challenges, solutions, reflections and recommendations.Philos Trans A Math Phys Eng Sci. 2022 Oct 3;380(2233):20210299. doi: 10.1098/rsta.2021.0299. Epub 2022 Aug 15. Philos Trans A Math Phys Eng Sci. 2022. PMID: 35965467 Free PMC article.
-
An analytical approach to evaluate the impact of age demographics in a pandemic.Stoch Environ Res Risk Assess. 2023 Jun 8:1-15. doi: 10.1007/s00477-023-02477-2. Online ahead of print. Stoch Environ Res Risk Assess. 2023. PMID: 37362847 Free PMC article.
-
Heterogeneity in the onwards transmission risk between local and imported cases affects practical estimates of the time-dependent reproduction number.Philos Trans A Math Phys Eng Sci. 2022 Oct 3;380(2233):20210308. doi: 10.1098/rsta.2021.0308. Epub 2022 Aug 15. Philos Trans A Math Phys Eng Sci. 2022. PMID: 35965464 Free PMC article.
-
Technical challenges of modelling real-life epidemics and examples of overcoming these.Philos Trans A Math Phys Eng Sci. 2022 Oct 3;380(2233):20220179. doi: 10.1098/rsta.2022.0179. Epub 2022 Aug 15. Philos Trans A Math Phys Eng Sci. 2022. PMID: 35965472 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

