Identification of key genes and infiltrating immune cells among acetaminophen-induced acute liver failure and HBV-associated acute liver failure
- PMID: 35965803
- PMCID: PMC9372688
- DOI: 10.21037/atm-22-2742
Identification of key genes and infiltrating immune cells among acetaminophen-induced acute liver failure and HBV-associated acute liver failure
Abstract
Background: Acute liver failure (ALF) is a life-threatening complication that is relatively uncommon. ALF causes severe hepatocyte damage and necrosis, which can lead to liver dysfunction and even multi-organ failure. A growing body of evidence suggests that immune cell infiltration and some abnormally expressed genes are associated with ALF development. However, in ALF, they have yet to be thoroughly investigated.
Methods: The Gene Expression Omnibus (GEO) database was used to obtain microarray datasets such as GSE74000, GSE120652, GSE38941, and GSE14668, which were then examined via GEO2R to determine differentially expressed genes (DEGs) associated with ALF. Metascape was employed to annotate the underlined genes using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. The mechanism of IGF1 in 2 different kinds of ALF including acetaminophen-induced ALF and hepatitis B virus (HBV)-induced ALF was studied using gene set enrichment analysis (GSEA). Next, immune cell infiltration was investigated and differentiated in ALF using CIBERSORT.
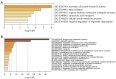
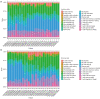
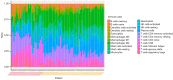
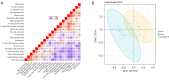
Results: Six genes (HAO2, IGF1, PLA2G7, SC5D, GNE, SLC1A1) were found to be abnormally expressed in the 2 distinct types of ALF i.e., acetaminophen-induced ALF and HBV-induced ALF. IGF1 was identified as a hub gene in ALF and was found to be associated with several developmental cascades including immune responses, inflammatory responses, and intracellular calcium homeostasis. Additionally, the number of CD4 naive T cells, CD8 T cells, and follicular helper T cells was increased in acetaminophen-induced ALF, whereas the number of activated NK cells, resting NK cells, and plasma cells was increased in HBV-induced ALF.
Conclusions: The present study determined a potential molecular target, namely IGF1, in acetaminophen-induced ALF and HBV-induced ALF, which may provide novel insights into the pathophysiology and management of ALF. Concurrently, the putative immunological pathways have been found.
Keywords: HBV-associated acute liver failure; IGF1; acetaminophen-induced acute liver failure; immune cell infiltration.
2022 Annals of Translational Medicine. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-2742/coif). The authors have no conflicts of interest to declare.
Figures





Similar articles
-
Bioinformatic identification of key genes and pathways that may be involved in the pathogenesis of HBV-associated acute liver failure.Genes Dis. 2018 Mar 2;5(4):349-357. doi: 10.1016/j.gendis.2018.02.005. eCollection 2018 Dec. Genes Dis. 2018. PMID: 30591937 Free PMC article.
-
Identification of Hub Genes and Potential Molecular Mechanisms in Patients with HBV-Associated Acute Liver Failure.Evol Bioinform Online. 2020 Oct 10;16:1176934320943901. doi: 10.1177/1176934320943901. eCollection 2020. Evol Bioinform Online. 2020. PMID: 33100826 Free PMC article.
-
Potential Biomarkers and Therapeutic Targets in Hepatitis B Virus-related Acute Liver Failure: Interplay of the Ferroptosis, Autophagy and Immune Responses.Int J Med Sci. 2025 Jan 21;22(4):806-818. doi: 10.7150/ijms.106360. eCollection 2025. Int J Med Sci. 2025. PMID: 39991755 Free PMC article.
-
Immune mechanisms in acetaminophen-induced acute liver failure.Hepatobiliary Surg Nutr. 2014 Dec;3(6):331-43. doi: 10.3978/j.issn.2304-3881.2014.11.01. Hepatobiliary Surg Nutr. 2014. PMID: 25568858 Free PMC article. Review.
-
Gc-globulin in liver disease.Dan Med Bull. 2008 Aug;55(3):131-46. Dan Med Bull. 2008. PMID: 19232164 Review.
Cited by
-
A Predictive Model to Evaluate the HbeAg Positivity of Chronic Hepatitis B Virus Patients in Clinics: A Cross-Sectional Study.Medicina (Kaunas). 2022 Sep 15;58(9):1279. doi: 10.3390/medicina58091279. Medicina (Kaunas). 2022. PMID: 36143956 Free PMC article.
-
Metabolic Health and Disease: A Role of Osteokines?Calcif Tissue Int. 2023 Jul;113(1):21-38. doi: 10.1007/s00223-023-01093-0. Epub 2023 May 17. Calcif Tissue Int. 2023. PMID: 37193929 Review.
-
Modifications of the GH Axis Reveal Unique Sexually Dimorphic Liver Signatures for Lcn13, Asns, Hamp2, Hao2, and Pgc1a.J Endocr Soc. 2024 Jan 31;8(3):bvae015. doi: 10.1210/jendso/bvae015. eCollection 2024 Jan 16. J Endocr Soc. 2024. PMID: 38370444 Free PMC article.
-
Hepatic antioxidant capacity, immune response, and glycolysis of Tibetan sheep in response to dietary soluble protein levels.Protoplasma. 2025 Sep;262(5):1183-1194. doi: 10.1007/s00709-025-02052-2. Epub 2025 Mar 19. Protoplasma. 2025. PMID: 40102302
-
Integrative Analyses of Genes of Pediatric Non-alcoholic Fatty Liver Disease Associated with Energy Metabolism.Dig Dis Sci. 2024 Dec;69(12):4373-4391. doi: 10.1007/s10620-024-08702-4. Epub 2024 Nov 4. Dig Dis Sci. 2024. PMID: 39496907 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
