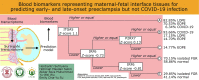
Blood biomarkers representing maternal-fetal interface tissues used to predict early-and late-onset preeclampsia but not COVID-19 infection
- PMID: 35966044
- PMCID: PMC9359600
- DOI: 10.1016/j.csbj.2022.08.011
Blood biomarkers representing maternal-fetal interface tissues used to predict early-and late-onset preeclampsia but not COVID-19 infection
Abstract
Background: A well-known blood biomarker (soluble fms-like tyrosinase-1 [sFLT-1]) for preeclampsia, i.e., a pregnancy disorder, was found to predict severe COVID-19, including in males. True biomarker may be masked by more-abrupt changes related to endothelial instead of placental dysfunction. This study aimed to identify blood biomarkers that represent maternal-fetal interface tissues for predicting preeclampsia but not COVID-19 infection.
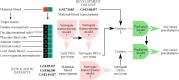
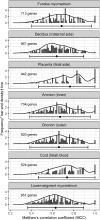
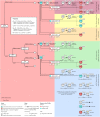
Methods: The surrogate transcriptome of tissues was determined by that in maternal blood, utilizing four datasets (n = 1354) which were collected before the COVID-19 pandemic. Applying machine learning, a preeclampsia prediction model was chosen between those using blood transcriptome (differentially expressed genes [DEGs]) and the blood-derived surrogate for tissues. We selected the best predictive model by the area under the receiver operating characteristic (AUROC) using a dataset for developing the model, and well-replicated in datasets both with and without an intervention. To identify eligible blood biomarkers that predicted any-onset preeclampsia from the datasets but that were not positive in the COVID-19 dataset (n = 47), we compared several methods of predictor discovery: (1) the best prediction model; (2) gene sets of standard pipelines; and (3) a validated gene set for predicting any-onset preeclampsia during the pandemic (n = 404). We chose the most predictive biomarkers from the best method with the significantly largest number of discoveries by a permutation test. The biological relevance was justified by exploring and reanalyzing low- and high-level, multiomics information.
Results: A prediction model using the surrogates developed for predicting any-onset preeclampsia (AUROC of 0.85, 95 % confidence interval [CI] 0.77 to 0.93) was the only that was well-replicated in an independent dataset with no intervention. No model was well-replicated in datasets with a vitamin D intervention. None of the blood biomarkers with high weights in the best model overlapped with blood DEGs. Blood biomarkers were transcripts of integrin-α5 (ITGA5), interferon regulatory factor-6 (IRF6), and P2X purinoreceptor-7 (P2RX7) from the prediction model, which was the only method that significantly discovered eligible blood biomarkers (n = 3/100 combinations, 3.0 %; P =.036). Most of the predicted events (73.70 %) among any-onset preeclampsia were cluster A as defined by ITGA5 (Z-score ≥ 1.1), but were only a minority (6.34 %) among positives in the COVID-19 dataset. The remaining were predicted events (26.30 %) among any-onset preeclampsia or those among COVID-19 infection (93.66 %) if IRF6 Z-score was ≥-0.73 (clusters B and C), in which none was the predicted events among either late-onset preeclampsia (LOPE) or COVID-19 infection if P2RX7 Z-score was <0.13 (cluster C). Greater proportions of predicted events among LOPE were cluster A (82.85 % vs 70.53 %) compared to early-onset preeclampsia (EOPE). The biological relevance by multiomics information explained the biomarker mechanism, polymicrobial infection in any-onset preeclampsia by ITGA5, viral co-infection in EOPE by ITGA5-IRF6, a shared prediction with COVID-19 infection by ITGA5-IRF6-P2RX7, and non-replicability in datasets with a vitamin D intervention by ITGA5.
Conclusions: In a model that predicts preeclampsia but not COVID-19 infection, the important predictors were genes in maternal blood that were not extremely expressed, including the proposed blood biomarkers. The predictive performance and biological relevance should be validated in future experiments.
Keywords: Biomarker; COVID-19; Machine learning; Preeclampsia; Transcriptome.
© 2022 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Similar articles
-
Lipidomic signatures in patients with early-onset and late-onset Preeclampsia.Metabolomics. 2024 Jun 16;20(4):65. doi: 10.1007/s11306-024-02134-x. Metabolomics. 2024. PMID: 38879866 Free PMC article.
-
Predicting preeclampsia in early pregnancy using clinical and laboratory data via machine learning model.BMC Med Inform Decis Mak. 2025 May 1;25(1):178. doi: 10.1186/s12911-025-02999-5. BMC Med Inform Decis Mak. 2025. PMID: 40312361 Free PMC article.
-
The feasibility of soluble Fms-Like Tyrosine kinase-1 (sFLT-1) and Placental Growth Factor (PlGF) ratio biomarker in predicting preeclampsia and adverse pregnancy outcomes among medium to high risk mothers in Kuala Lumpur, Malaysia.PLoS One. 2022 Mar 11;17(3):e0265080. doi: 10.1371/journal.pone.0265080. eCollection 2022. PLoS One. 2022. PMID: 35275947 Free PMC article.
-
Combining Biomarkers to Predict Pregnancy Complications and Redefine Preeclampsia: The Angiogenic-Placental Syndrome.Hypertension. 2020 Apr;75(4):918-926. doi: 10.1161/HYPERTENSIONAHA.119.13763. Epub 2020 Feb 17. Hypertension. 2020. PMID: 32063058 Free PMC article. Review.
-
Oxidative stress and mitochondrial dysfunction in early-onset and late-onset preeclampsia.Biochim Biophys Acta Mol Basis Dis. 2020 Dec 1;1866(12):165961. doi: 10.1016/j.bbadis.2020.165961. Epub 2020 Sep 8. Biochim Biophys Acta Mol Basis Dis. 2020. PMID: 32916282 Review.
References
-
- Espino Y.S.S., Martinez-Portilla R.J., Torres-Torres J., Solis-Paredes J.M., Estrada-Gutierrez G., et al. Novel ratio soluble fms-like tyrosine kinase-1/angiotensin-ii (sflt-1/ang-ii) in pregnant women is associated with critical illness in covid-19. Viruses. 2021;13 doi: 10.3390/v13101906. - DOI - PMC - PubMed
-
- Dupont V., Kanagaratnam L., Goury A., Poitevin G., Bard M., et al. Excess soluble fms-like tyrosine kinase 1 correlates with endothelial dysfunction and organ failure in critically ill coronavirus disease 2019 patients. Clin Infect Dis. 2021;72:1834–1837. doi: 10.1093/cid/ciaa1007. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
