5-Hydroxymethylcytosine profiles in plasma cell-free DNA reflect molecular characteristics of diabetic kidney disease
- PMID: 35966076
- PMCID: PMC9372268
- DOI: 10.3389/fendo.2022.910907
5-Hydroxymethylcytosine profiles in plasma cell-free DNA reflect molecular characteristics of diabetic kidney disease
Abstract
Background: Diabetic kidney disease (DKD), one of the main complications of diabetes mellitus (DM), has become a frequent cause of end-stage renal disease. A clinically convenient, non-invasive approach for monitoring the development of DKD would benefit the overall life quality of patients with DM and contribute to lower medical burdens through promoting preventive interventions.
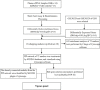
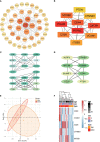
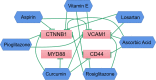
Methods: We utilized 5hmC-Seal to profile genome-wide 5-hydroxymethylcytosines in plasma cell-free DNA (cfDNA). Candidate genes were identified by intersecting the differentially hydroxymethylated genes and differentially expressed genes from the GSE30528 and GSE30529. Then, a protein interaction network was constructed for the candidate genes, and the hub genes were identified by the MCODE and cytoHubba algorithm. The correlation analysis between the hydroxymethylation level of the hub genes and estimated glomerular filtration rate (eGFR) was carried out. Finally, we demonstrated differences in expression levels of the protein was verified by constructing a mouse model of DKD. In addition, we constructed a network of interactions between drugs and hub genes using the Comparative Toxicogenomics Database.
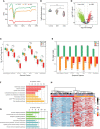
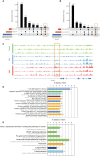
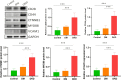
Results: This study found that there were significant differences in the overall distribution of 5hmC in plasma of patients with DKD, and an alteration of hydroxymethylation levels in genomic regions involved in inflammatory pathways which participate in the immune response. The final 5 hub genes, including (CTNNB1, MYD88, CD28, VCAM1, CD44) were confirmed. Further analysis indicated that this 5-gene signature showed a good capacity to distinguish between DKD and DM, and was found that protein levels were increased in renal tissue of DKD mice. Correlation analysis indicated that the hydroxymethylation level of 5 hub genes were nagatively correlated with eGFR. Toxicogenomics analysis showed that a variety of drugs for the treatment of DKD can reduce the expression levels of 4 hub genes (CD44, MYD88, VCAM1, CTNNB1).
Conclusions: The 5hmC-Seal assay was successfully applied to the plasma cfDNA samples from a cohort of DM patients with or without DKD. Altered 5hmC signatures indicate that 5hmC-Seal has the potential to be a non-invasive epigenetic tool for monitoring the development of DKD and it provides new insight for the future molecularly targeted anti-inflammation therapeutic strategies of DKD.
Keywords: 5-hydroxymethylcytosine 5-; Epigenetics; biomarker; cell-free DNDNA; diabetic kidney disease.
Copyright © 2022 Chu, Bi, He, Ma, Wan, Wang, Zhang, Zheng, Yang, Du, Maimaiti, Biekedawulaiti, Duolikun, Chen, Chen, Li, Tie and Lin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Thomas MC, Macisaac RJ, Jerums G, Weekes A, Moran J, Shaw JE, et al. Nonalbuminuric renal impairment in type 2 diabetic patients and in the general population (national evaluation of the frequency of renal impairment cO-existing with NIDDM [NEFRON] 11). Diabetes Care (2009) 32:1497–502. doi: 10.2337/dc08-2186 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

