Identification of a pyroptosis related gene signature for predicting prognosis and estimating tumor immune microenvironment in bladder cancer
- PMID: 35966309
- PMCID: PMC9372197
- DOI: 10.21037/tcr-22-177
Identification of a pyroptosis related gene signature for predicting prognosis and estimating tumor immune microenvironment in bladder cancer
Abstract
Background: Recently, there are growing evidence indicated that pyroptosis play a critical role in the incidence of many diseases. Here, we aimed to identify the specific function and prognosis predictive of pyroptosis-related genes (PRGs) in bladder cancer (BLCA) patients.
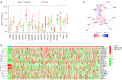
Methods: The gene expression and corresponding clinical data of BLCA patients were obtained from The Cancer Genome Atlas (TCGA), and the expression level of PRGs was identified between normal and tumor tissues. Furthermore, univariate Cox regression was conducted to filter the PRGs related to overall survival, and LASSO Cox regression was subsequently conducted to establish the PRGs risk model. Besides, the correlation of risk score with patients' clinical features, tumor mutational burden (TMB) as well as tumor microenvironment (TME) was also investigated.
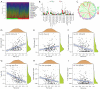
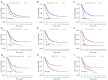
Results: A total of 6 PRGs was used to establish the risk prognostic model. According the median value of risk score, the patients were classified into low- and high-risk subgroup. Kaplan-Meier survival analysis revealed that the BLCA patients in low-risk group exhibited a better survival prognosis compared with high-risk group. More important, after adjusting for age, gender, tumor grade, and clinical stage, the risk score resulted as an independent factor affecting the clinical prognosis of BLCA patients. In addition, the PRGs risk signature was also correlated with immune cell infiltration, TMB and TME.
Conclusions: The present study offered a novel PRGs risk model to access the clinical prognosis of BLCA and provided new insight for future study to improve overall survival and responses to cancer therapy targeting pyroptosis.
Keywords: Bladder cancer (BLCA); biomarker; prognosis; pyroptosis.
2022 Translational Cancer Research. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-22-177/coif). The authors have no conflicts of interest to declare.
Figures






References
LinkOut - more resources
Full Text Sources
