Comparative Genomic Analysis of Seven Vibrio alginolyticus Strains Isolated From Shrimp Larviculture Water With Emphasis on Chitin Utilization
- PMID: 35966654
- PMCID: PMC9364117
- DOI: 10.3389/fmicb.2022.925747
Comparative Genomic Analysis of Seven Vibrio alginolyticus Strains Isolated From Shrimp Larviculture Water With Emphasis on Chitin Utilization
Abstract
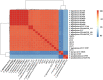
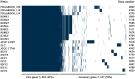
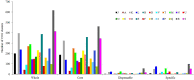
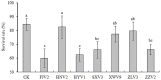
The opportunistic pathogen Vibrio alginolyticus is gaining attention because of its disease-causing risks to aquatic animals and humans. In this study, seven Vibrio strains isolated from different shrimp hatcheries in Southeast China were subjected to genome sequencing and subsequent comparative analysis to explore their intricate relationships with shrimp aquaculture. The seven isolates had an average nucleotide identity of ≥ 98.3% with other known V. alginolyticus strains. The species V. alginolyticus had an open pan-genome, with the addition of ≥ 161 novel genes following each new genome for seven isolates and 14 publicly available V. alginolyticus strains. The percentages of core genes of the seven strains were up to 83.1-87.5%, indicating highly conserved functions, such as chitin utilization. Further, a total of 14 core genes involved in the chitin degradation pathway were detected on the seven genomes with a single copy, 12 of which had undergone significant purifying selection (dN/dS < 1). Moreover, the seven strains could utilize chitin as the sole carbon-nitrogen source. In contrast, mobile genetic elements (MGEs) were identified in seven strains, including plasmids, prophages, and genomic islands, which mainly encoded accessory genes annotated as hypothetical proteins. The infection experiment showed that four of the seven strains might be pathogenic because the survival rates of Litopenaeus vannamei postlarvae were significantly reduced (P < 0.05) when compared to the control. However, no obvious correlation was noted between the number of putative virulence factors and toxic effects of the seven strains. Collectively, the persistence of V. alginolyticus in various aquatic environments may be attributed to its high genomic plasticity via the acquisition of novel genes by various MGEs. In view of the strong capability of chitin utilization by diverse vibrios, the timely removal of massive chitin-rich materials thoroughly in shrimp culture systems may be a key strategy to inhibit proliferation of vibrios and subsequent infection of shrimp. In addition, transcontinental transfer of potentially pathogenic V. alginolyticus strains should receive great attention to avoid vibriosis.
Keywords: Vibrio alginolyticus; chitin utilization; mobile genetic element; pan-genome; shrimp larviculture.
Copyright © 2022 Xue, Huang, Xue, He, Liang, Liang, Liu and Wen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Comparative Genomic Analysis of Shrimp-Pathogenic Vibrio parahaemolyticus LC and Intraspecific Strains with Emphasis on Virulent Factors of Mobile Genetic Elements.Microorganisms. 2023 Nov 11;11(11):2752. doi: 10.3390/microorganisms11112752. Microorganisms. 2023. PMID: 38004763 Free PMC article.
-
An in vitro and in vivo assessment of the potential of Vibrio spp. as probiotics for the Pacific white shrimp, Litopenaeus vannamei.J Appl Microbiol. 2010 Oct;109(4):1177-87. doi: 10.1111/j.1365-2672.2010.04743.x. Epub 2010 Aug 19. J Appl Microbiol. 2010. PMID: 20477892
-
Genomic variation among closely related Vibrio alginolyticus strains is located on mobile genetic elements.BMC Genomics. 2020 May 11;21(1):354. doi: 10.1186/s12864-020-6735-5. BMC Genomics. 2020. PMID: 32393168 Free PMC article.
-
The protective effect of chitin and chitosan against Vibrio alginolyticus in white shrimp Litopenaeus vannamei.Fish Shellfish Immunol. 2005 Sep;19(3):191-204. doi: 10.1016/j.fsi.2004.11.003. Fish Shellfish Immunol. 2005. PMID: 15820121
-
Vibrios associated with Litopenaeus vannamei larvae, postlarvae, broodstock, and hatchery probionts.Appl Environ Microbiol. 1999 Jun;65(6):2592-7. doi: 10.1128/AEM.65.6.2592-2597.1999. Appl Environ Microbiol. 1999. PMID: 10347048 Free PMC article.
Cited by
-
Insights into Chitin-Degradation Potential of Shewanella khirikhana JW44 with Emphasis on Characterization and Function of a Chitinase Gene SkChi65.Microorganisms. 2024 Apr 11;12(4):774. doi: 10.3390/microorganisms12040774. Microorganisms. 2024. PMID: 38674717 Free PMC article.
-
Comparative Genomic Analysis of Shrimp-Pathogenic Vibrio parahaemolyticus LC and Intraspecific Strains with Emphasis on Virulent Factors of Mobile Genetic Elements.Microorganisms. 2023 Nov 11;11(11):2752. doi: 10.3390/microorganisms11112752. Microorganisms. 2023. PMID: 38004763 Free PMC article.
References
-
- Abdul Hannan M. D., Mahbubur Rahman M. D., Nurunnabi Mondal M. D., Suzan Chandra D., Chowdhury G., Tofazzal Islam M. D. (2019). Molecular identification of Vibrio alginolyticus causing vibriosis in shrimp an its herbal remedy. Pol. J. Microbiol. 68, 429–438. 10.33073/pjm-2019-042 - DOI - PMC - PubMed
-
- Bachand P. T., Tallmana J. J., Powers N. C., Woods M., Azadania D. N., Zimba P. V., et al. . (2020). Genomic identification and characterization of co-occurring Harveyi clade species following a vibriosis outbreak in Pacific white shrimp, Penaeus (litopenaeus) vannamei. Aquaculture 518:734628. 10.1016/j.aquaculture.2019.734628 - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

