The effect of environment on intestinal microbial diversity of Panthera animals may exceed genetic relationship
- PMID: 35966667
- PMCID: PMC9366613
- DOI: 10.3389/fmicb.2022.938900
The effect of environment on intestinal microbial diversity of Panthera animals may exceed genetic relationship
Abstract
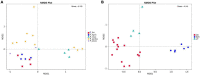
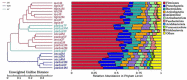
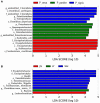
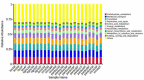
Intestinal microbes are important symbiotes in the gastrointestinal tract of mammals, which are affected by food, environment, climate, genetics, and other factors. The gut microbiota of felines has been partially studied, but a comprehensive comparison of the gut microbiota of Panthera species was less reported. In this study, we compared the gut microbial composition and diversity of five species of Panthera (Panthera tigris, Panthera leo, Panthera onca, Panthera pardus, and Panthera uncia) by 16S ribosomal RNA (rRNA) amplicon sequencing. The results showed that Firmicutes was the most abundant phylum among all the Panthera species, followed by Actinobacteria, Fusobacteria, Bacteroidetes, Proteobacteria, Acidobacteria, Verrucomicrobia, Gemmatimonadetes, and Euryarchaeota. There were significant differences in observed species of fecal microbiota among different Panthera animals (P < 0.05), indicating that there is species specificity among Panthera fecal microbiota. When the samples were further grouped according to sampling locations, the comparison of the alpha diversity index between groups and beta diversity analysis showed that there were significant differences in the fecal microflora of animals from different sampling locations. Cluster analysis showed that fecal microbes of animals from the same sampling location were clustered, while gut microbes of animals of the same species, but from different sampling locations, were separated. These results indicate that environment may have more influence on mammals' fecal microbial diversity than genetic relationships.
Keywords: Panthera animals; diversity analysis; genetic relationship; gut microbiota; sampling environment.
Copyright © 2022 Chen, Xu, Sun, Li, Wang, Gao, Gao and Shi.
Conflict of interest statement
LC was employed by the Qufu Normal University. DX, MS, SW, ZG, and YS are studying at Qufu Normal University. YL and YG were employed by Jinan Wildlife Park.
Figures
Similar articles
-
Habitat shapes the gut microbiome diversity of Malayan tigers (Panthera tigris jacksoni) as revealed through metabarcoding 16S rRNA profiling.World J Microbiol Biotechnol. 2024 Feb 28;40(4):111. doi: 10.1007/s11274-023-03868-x. World J Microbiol Biotechnol. 2024. PMID: 38416247
-
Comparison of the fecal microbiota of two free-ranging Chinese subspecies of the leopard (Panthera pardus) using high-throughput sequencing.PeerJ. 2019 Mar 28;7:e6684. doi: 10.7717/peerj.6684. eCollection 2019. PeerJ. 2019. PMID: 30944781 Free PMC article.
-
Gut microbiota and fecal metabolites in captive and wild North China leopard (Panthera pardus japonensis) by comparsion using 16 s rRNA gene sequencing and LC/MS-based metabolomics.BMC Vet Res. 2020 Sep 29;16(1):363. doi: 10.1186/s12917-020-02583-1. BMC Vet Res. 2020. PMID: 32993639 Free PMC article.
-
Colonization and Development of the Fecal Microflora of South China Tiger Cubs (Panthera tigris amoyensis) by Sequencing of the 16S rRNA Gene.Microb Physiol. 2022;32(1-2):18-29. doi: 10.1159/000518395. Epub 2021 Sep 14. Microb Physiol. 2022. PMID: 34535588
-
Mitogenomic analysis of the genus Panthera.Sci China Life Sci. 2011 Oct;54(10):917-30. doi: 10.1007/s11427-011-4219-1. Epub 2011 Oct 29. Sci China Life Sci. 2011. PMID: 22038004
Cited by
-
High-throughput DNA metabarcoding for determining the gut microbiome of captive critically endangered Malayan tiger (Pantheratigrisjacksoni) during fasting.Biodivers Data J. 2023 Sep 5;11:e104757. doi: 10.3897/BDJ.11.e104757. eCollection 2023. Biodivers Data J. 2023. PMID: 37711366 Free PMC article.
-
Alteration of the gut microbial composition of critically endangered Malayan tigers (Panthera tigris jacksoni) in captivity during enrichment phase.Mol Biol Rep. 2024 Jun 14;51(1):742. doi: 10.1007/s11033-024-09642-y. Mol Biol Rep. 2024. PMID: 38874703
-
Habitat shapes the gut microbiome diversity of Malayan tigers (Panthera tigris jacksoni) as revealed through metabarcoding 16S rRNA profiling.World J Microbiol Biotechnol. 2024 Feb 28;40(4):111. doi: 10.1007/s11274-023-03868-x. World J Microbiol Biotechnol. 2024. PMID: 38416247
-
Gut microbiome of captive wolves is more similar to domestic dogs than wild wolves indicated by metagenomics study.Front Microbiol. 2022 Nov 1;13:1027188. doi: 10.3389/fmicb.2022.1027188. eCollection 2022. Front Microbiol. 2022. PMID: 36386659 Free PMC article.
References
-
- Dexter F. (2013). Wilcoxon-Mann-Whitney test used for data that are not normally distributed. Anesth. Analg. 117 537–538. - PubMed
LinkOut - more resources
Full Text Sources

