The ABCT31 Transporter Regulates the Export System of Phenylacetic Acid as a Side-Chain Precursor of Penicillin G in Monascus ruber M7
- PMID: 35966689
- PMCID: PMC9370074
- DOI: 10.3389/fmicb.2022.915721
The ABCT31 Transporter Regulates the Export System of Phenylacetic Acid as a Side-Chain Precursor of Penicillin G in Monascus ruber M7
Abstract
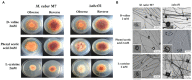
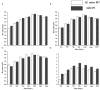
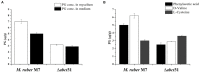
The biosynthesis of penicillin G (PG) is compartmentalized, and the transportation of the end and intermediate products, and substrates (precursors) such as L-cysteine (L-Cys), L-valine (L-Val) and phenylacetic acid (PAA) requires traversing membrane barriers. However, the transportation system of PAA as a side chain of PG are unclear yet. To discover ABC transporters (ABCTs) involved in the transportation of PAA, the expression levels of 38 ABCT genes in the genome of Monascus ruber M7, culturing with and without PAA, were examined, and found that one abct gene, namely abct31, was considerably up-regulated with PAA, indicating that abct31 may be relative with PAA transportation. Furthermore the disruption of abct31 was carried out, and the effects of two PG substrate's amino acids (L-Cys and L-Val), PAA and some other weak acids on the morphologies and production of secondary metabolites (SMs) of Δabct31 and M. ruber M7, were performed through feeding experiments. The results revealed that L-Cys, L-Val and PAA substantially impacted the morphologies and SMs production of Δabct31 and M. ruber M7. The UPLC-MS/MS analysis findings demonstrated that Δabct31 did not interrupt the synthesis of PG in M. ruber M7. According to the results, it suggests that abct31 is involved in the resistance and detoxification of the weak acids, including the PAA in M. ruber M7.
Keywords: ABC transporter; amino acid; phenylacetic acid; secondary metabolites; weak acid.
Copyright © 2022 Ramzan, Virk and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures









References
-
- Al Shawi A., Rasul A., Khan M., Iqbal F., Tonghui M. (2011). Eupatilin: a flavonoid compound isolated from the artemisia plant, induces apoptosis and G2/M phase cell cycle arrest in human melanoma A375 cells. African J. Pharm. Pharmacol. 5, 582–588. 10.5897/AJPP11.079 - DOI
-
- Ávalos J., Díaz-Sánchez V., García-Martínez, Jorge Castrillo M., Ruger-Herreros M., Limón M. C. (2014). Biosynthesis and molecular genetics of fungal secondary metabolites. Fungal Biol. 2, 67–79. 10.1007/978-1-4939-1191-2 - DOI
LinkOut - more resources
Full Text Sources

