Spatio-Temporal Variation in the Phyllospheric Microbial Biodiversity of Alternaria Alternata-Infected Tobacco Foliage
- PMID: 35966692
- PMCID: PMC9370072
- DOI: 10.3389/fmicb.2022.920109
Spatio-Temporal Variation in the Phyllospheric Microbial Biodiversity of Alternaria Alternata-Infected Tobacco Foliage
Abstract
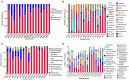
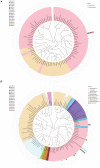
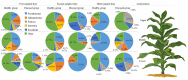
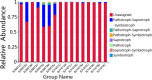
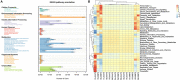
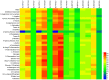
Phyllospheric microbial composition of tobacco (Nicotiana tabacum L.) is contingent upon certain factors, such as the growth stage of the plant, leaf position, and cultivar and its geographical location, which influence, either directly or indirectly, the growth, overall health, and production of the tobacco plant. To better understand the spatiotemporal variation of the community and the divergence of phyllospheric microflora, procured from healthy and diseased tobacco leaves infected by Alternaria alternata, the current study employed microbe culturing, high-throughput technique, and BIOLOG ECO. Microbe culturing resulted in the isolation of 153 culturable fungal isolates belonging to 33 genera and 99 bacterial isolates belonging to 15 genera. High-throughput sequencing revealed that the phyllosphere of tobacco was dominantly colonized by Ascomycota and Proteobacteria, whereas, the most abundant fungal and bacterial genera were Alternaria and Pseudomonas. The relative abundance of Alternaria increased in the upper and middle healthy groups from the first collection time to the third, whereas, the relative abundance of Pseudomonas, Sphingomonas, and Methylobacterium from the same positions increased during gradual leaf aging. Non-metric multi-dimensional scaling (NMDs) showed clustering of fungal communities in healthy samples, while bacterial communities of all diseased and healthy groups were found scattered. FUNGuild analysis, from the first collection stage to the third one in both groups, indicated an increase in the relative abundance of Pathotroph-Saprotroph, Pathotroph-Saprotroph-Symbiotroph, and Pathotroph-Symbiotroph. Inclusive of all samples, as per the PICRUSt analysis, the predominant pathway was metabolism function accounting for 50.03%. The average values of omnilog units (OUs) showed relatively higher utilization rates of carbon sources by the microbial flora of healthy leaves. According to the analysis of genus abundances, leaf growth and leaf position were the important drivers of change in structuring the microbial communities. The current findings revealed the complex ecological dynamics that occur in the phyllospheric microbial communities over the course of a spatiotemporal varying environment with the development of tobacco brown spots, highlighting the importance of community succession.
Keywords: Alternaria alternata; biolog-eco; high-throughput sequencing; microbial community; microbial functional diversity; tobacco brown spot.
Copyright © 2022 Dai, Wu, Wang, Li, Cai, Li, Wang, Sehar and Shamsi.
Conflict of interest statement
Y-fD was employed by Bijie Tobacco Company. J-xL was employed by Guizhou Tobacco Company of CNTC, China National Tobacco Corporation. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Effect of disease severity on the structure and diversity of the phyllosphere microbial community in tobacco.Front Microbiol. 2023 Jan 4;13:1081576. doi: 10.3389/fmicb.2022.1081576. eCollection 2022. Front Microbiol. 2023. PMID: 36687583 Free PMC article.
-
Response of microbial communities in the phyllosphere ecosystem of tobacco exposed to the broad-spectrum copper hydroxide.Front Microbiol. 2023 Sep 28;14:1229294. doi: 10.3389/fmicb.2023.1229294. eCollection 2023. Front Microbiol. 2023. PMID: 37840714 Free PMC article.
-
Variations in leaf phyllosphere microbial communities and development of tobacco brown spot before and after fungicide application.Front Microbiol. 2022 Nov 17;13:1068158. doi: 10.3389/fmicb.2022.1068158. eCollection 2022. Front Microbiol. 2022. PMID: 36466663 Free PMC article.
-
Incentives for preventing smoking in children and adolescents.Cochrane Database Syst Rev. 2017 Jun 6;6(6):CD008645. doi: 10.1002/14651858.CD008645.pub3. Cochrane Database Syst Rev. 2017. PMID: 28585288 Free PMC article.
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article.
Cited by
-
Characteristics of Epicoccum latusicollum as revealed by genomic and metabolic phenomic analysis, the causal agent of tobacco Epicoccus leaf spot.Front Plant Sci. 2023 Aug 24;14:1199956. doi: 10.3389/fpls.2023.1199956. eCollection 2023. Front Plant Sci. 2023. PMID: 37828924 Free PMC article.
-
Effect of disease severity on the structure and diversity of the phyllosphere microbial community in tobacco.Front Microbiol. 2023 Jan 4;13:1081576. doi: 10.3389/fmicb.2022.1081576. eCollection 2022. Front Microbiol. 2023. PMID: 36687583 Free PMC article.
-
Response of microbial communities in the tobacco phyllosphere under the stress of validamycin.Front Microbiol. 2024 Jan 18;14:1328179. doi: 10.3389/fmicb.2023.1328179. eCollection 2023. Front Microbiol. 2024. PMID: 38304858 Free PMC article.
-
Optimizing greenhouse microclimate for plant pathology: challenges and cooling solutions for pathogen control in arid regions.Front Plant Sci. 2025 Feb 6;16:1492760. doi: 10.3389/fpls.2025.1492760. eCollection 2025. Front Plant Sci. 2025. PMID: 39980477 Free PMC article. Review.
-
Pan-metagenome reveals the abiotic stress resistome of cigar tobacco phyllosphere microbiome.Front Plant Sci. 2023 Dec 21;14:1248476. doi: 10.3389/fpls.2023.1248476. eCollection 2023. Front Plant Sci. 2023. PMID: 38179476 Free PMC article.
References
-
- Almasoud J., McArdle L., Henne R., Mathews K., O'Connor Í., McLoughlin L. E., et al. (2019). Draft genome sequence of a red basidiomycete yeast, symmetrospora coprosmae strain UCD350, isolated from soil in Ireland. Microbiol. Resourc. Announc. 8, e01256–e01219. 10.1128/MRA.01256-19 - DOI - PMC - PubMed
-
- Attitalla I. H., Al-Ani L. K. T., Nasib M. A., Balal I. A. A., Zakaria M., El-maraghy S. S. M., Rezaul K. S. M. (2010). Screening of fungi associated with commercial grains and animal feeds in Al-Bayda Governorate, Libya. World Appl. Sci. J. 9, 746–756. Available online at: https://www.researchgate.net/publication/280253933_Screening_of_Fungi_As...
-
- Barcellos F. G., Menna P., da Silva Batista J. S., Hungria M. (2007). Evidence of Horizontal Transfer of Symbiotic Genes from a Bradyrhizobium japonicum Inoculant Strain to Indigenous Diazotrophs Sinorhizobium (Ensifer) fredii and Bradyrhizobium elkanii in a Brazilian Savannah Soil. Applied and Environ. Microbiol. 73, 2635–2643. 10.1128/aem.01823-06 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

