Molecular and biological characterization of hepatitis B virus subgenotype F1b clusters: Unraveling its role in hepatocarcinogenesis
- PMID: 35966715
- PMCID: PMC9363773
- DOI: 10.3389/fmicb.2022.946703
Molecular and biological characterization of hepatitis B virus subgenotype F1b clusters: Unraveling its role in hepatocarcinogenesis
Abstract
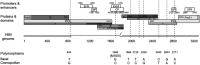
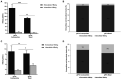
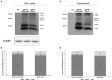
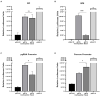
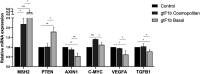
Hepatitis B virus (HBV) subgenotype F1b infection has been associated with the early occurrence of hepatocellular carcinoma in chronically infected patients from Alaska and Peru. In Argentina, however, despite the high prevalence of subgenotype F1b infection, this relationship has not been described. To unravel the observed differences in the progression of the infection, an in-depth molecular and biological characterization of the subgenotype F1b was performed. Phylogenetic analysis of subgenotype F1b full-length genomes revealed the existence of two highly supported clusters. One of the clusters, designated as gtF1b Basal included sequences mostly from Alaska, Peru and Chile, while the other, called gtF1b Cosmopolitan, contained samples mainly from Argentina and Chile. The clusters were characterized by a differential signature pattern of eight nucleotides distributed throughout the genome. In vitro characterization of representative clones from each cluster revealed major differences in viral RNA levels, virion secretion, antigen expression levels, as well as in the localization of the antigens. Interestingly, a differential regulation in the expression of genes associated with tumorigenesis was also identified. In conclusion, this study provides new insights into the molecular and biological characteristics of the subgenotype F1b clusters and contributes to unravel the different clinical outcomes of subgenotype F1b chronic infections.
Keywords: characterization; clusters; hepatitis B virus; hepatocarcinogenesis; subgenotype F1b.
Copyright © 2022 Elizalde, Mojsiejczuk, Speroni, Bouzas, Tadey, Mammana, Campos and Flichman.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Bouchard M. J., Puro R. J., Wang L., Schneider R. J. (2003). Activation and inhibition of cellular calcium and tyrosine kinase signaling pathways identify targets of the HBx protein involved in hepatitis B virus replication. J. Virol. 77, 7713–7719. doi: 10.1128/jvi.77.14.7713-7719.2003, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

