Whole genome sequencing of the multidrug-resistant Chryseobacterium indologenes isolated from a patient in Brazil
- PMID: 35966843
- PMCID: PMC9366087
- DOI: 10.3389/fmed.2022.931379
Whole genome sequencing of the multidrug-resistant Chryseobacterium indologenes isolated from a patient in Brazil
Abstract
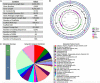
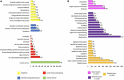
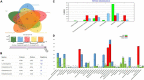
Chryseobacterium indologenes is a non-glucose-fermenting Gram-negative bacillus. This emerging multidrug resistant opportunistic nosocomial pathogen can cause severe infections in neonates and immunocompromised patients. This study aimed to present the first detailed draft genome sequence of a multidrug-resistant C. indologenes strain isolated from the cerebrospinal fluid of an infant hospitalized at the Neonatal Intensive Care Unit of Brazilian Tertiary Hospital. We first analyzed the susceptibility of C. indologenes strain to different antibiotics using the VITEK 2 system. The strain demonstrated an outstanding resistance to all the antibiotic classes tested, including β-lactams, aminoglycosides, glycylcycline, and polymyxin. Next, C. indologenes was whole-genome-sequenced, annotated using Prokka and Rapid Annotation using Subsystems Technology (RAST), and screened for orthologous groups (EggNOG), gene ontology (GO), resistance genes, virulence genes, and mobile genetic elements using different software tools. The draft genome contained one circular chromosome of 4,836,765 bp with 37.32% GC content. The genomic features of the chromosome present numerous genes related to cellular processes that are essential to bacteria. The MDR C. indologenes revealed the presence of genes that corresponded to the resistance phenotypes, including genes to β-lactamases (bla IND-13, bla CIA-3, bla TEM-116, bla OXA-209, bla VEB-15), quinolone (mcbG), tigecycline (tet(X6)), and genes encoding efflux pumps which confer resistance to aminoglycosides (RanA/RanB), and colistin (HlyD/TolC). Amino acid substitutions related to quinolone resistance were observed in GyrA (S83Y) and GyrB (L425I and K473R). A mutation that may play a role in the development of colistin resistance was detected in lpxA (G68D). Chryseobacterium indologenes isolate harbored 19 virulence factors, most of which were involved in infection pathways. We identified 13 Genomic Islands (GIs) and some elements associated with one integrative and conjugative element (ICEs). Other elements linked to mobile genetic elements (MGEs), such as insertion sequence (ISEIsp1), transposon (Tn5393), and integron (In31), were also present in the C. indologenes genome. Although plasmids were not detected, a ColRNAI replicon type and the most resistance genes detected in singletons were identified in unaligned scaffolds. We provided a wide range of information toward the understanding of the genomic diversity of C. indologenes, which can contribute to controlling the evolution and dissemination of this pathogen in healthcare settings.
Keywords: Chryseobacterium indologenes; metabolic features; mobile genetic elements; neonatal intensive care unit; virulence and resistance genes; whole-genome sequencing.
Copyright © 2022 Damas, Ferreira, Campanini, Soares, Campos, Laprega, Soares da Costa, Freire, Pitondo-Silva, Cerdeira, Cunha and Pranchevicius.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Whole genome sequencing uncovers a novel IND-16 metallo-β-lactamase from an extensively drug-resistant Chryseobacterium indologenes strain J31.Gut Pathog. 2016 Oct 21;8:47. doi: 10.1186/s13099-016-0130-4. eCollection 2016. Gut Pathog. 2016. PMID: 27785154 Free PMC article.
-
Brevundimonas brasiliensis sp. nov.: a New Multidrug-Resistant Species Isolated from a Patient in Brazil.Microbiol Spectr. 2023 Jun 15;11(3):e0441522. doi: 10.1128/spectrum.04415-22. Epub 2023 Apr 17. Microbiol Spectr. 2023. PMID: 37067439 Free PMC article.
-
Whole genome sequencing for deciphering the resistome of Chryseobacterium indologenes, an emerging multidrug-resistant bacterium isolated from a cystic fibrosis patient in Marseille, France.New Microbes New Infect. 2016 Apr 1;12:35-42. doi: 10.1016/j.nmni.2016.03.006. eCollection 2016 Jul. New Microbes New Infect. 2016. PMID: 27222716 Free PMC article.
-
A case of hospital-acquired pneumonia associated with Chryseobacterium indologenes infection in a patient with HIV infection and review of the literature.AIDS Res Ther. 2025 May 23;22(1):53. doi: 10.1186/s12981-025-00749-1. AIDS Res Ther. 2025. PMID: 40410755 Free PMC article. Review.
-
Six cases during 2012-2015 and literature review of Chryseobacterium indologenes infections in pediatric patients.Can J Microbiol. 2016 Oct;62(10):812-819. doi: 10.1139/cjm-2015-0800. Epub 2016 May 17. Can J Microbiol. 2016. PMID: 27397741 Review.
Cited by
-
Novel Antibiotic Resistance Genes Identified by Functional Gene Library Screening in Stenotrophomonas maltophilia and Chryseobacterium spp. Bacteria of Soil Origin.Int J Mol Sci. 2023 Mar 23;24(7):6037. doi: 10.3390/ijms24076037. Int J Mol Sci. 2023. PMID: 37047008 Free PMC article.
-
The tigecycline resistance mechanisms in Gram-negative bacilli.Front Cell Infect Microbiol. 2024 Nov 20;14:1471469. doi: 10.3389/fcimb.2024.1471469. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39635040 Free PMC article. Review.
-
Rarely Encountered Gram-Negative Rods and Lung Transplant Recipients: A Narrative Review.Microorganisms. 2023 May 31;11(6):1468. doi: 10.3390/microorganisms11061468. Microorganisms. 2023. PMID: 37374970 Free PMC article. Review.
-
First report of coexistence of blaKPC-2 and blaNDM-1 in carbapenem-resistant clinical isolates of Klebsiella aerogenes in Brazil.Front Microbiol. 2024 Feb 14;15:1352851. doi: 10.3389/fmicb.2024.1352851. eCollection 2024. Front Microbiol. 2024. PMID: 38426065 Free PMC article.
-
Two colistin resistance-producing Aeromonas strains, isolated from coastal waters in Zhejiang, China: characteristics, multi-drug resistance and pathogenicity.Front Microbiol. 2024 Jul 31;15:1401802. doi: 10.3389/fmicb.2024.1401802. eCollection 2024. Front Microbiol. 2024. PMID: 39144207 Free PMC article.
References
-
- Vandamme P, Bernadet J-F, Segers P, Kersters K, Holmes B. New perspectives in the classification of the flavobacteria: description of Chryseobacterium gen. nov., Bergeyella gen. nov., and Empedobacter nom. rev. Int J Syst Bacteriol. (1994) 44:827–31. 10.1099/00207713-44-4-827 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

