Full-length 16S rRNA amplicon sequencing reveals the variation of epibiotic microbiota associated with two shrimp species of Alvinocarididae: possibly co-determined by environmental heterogeneity and specific recognition of hosts
- PMID: 35966925
- PMCID: PMC9368993
- DOI: 10.7717/peerj.13758
Full-length 16S rRNA amplicon sequencing reveals the variation of epibiotic microbiota associated with two shrimp species of Alvinocarididae: possibly co-determined by environmental heterogeneity and specific recognition of hosts
Abstract
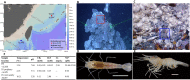
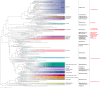
Shrimps of the family Alvinocarididae, endemic species to deep sea chemosynthetic ecosystems, harbor epibiotic microbes on gills which probably play important roles in the survival of the shrimps. Among them, Alvinocaris longirostris and Shinkaicaris leurokolos occupy different ecological niches within the same hydrothermal vent in Okinawa Trough, and A. longirostris also exists in a methane seep of the South China Sea. In this study, full-length 16S rRNA sequences of the gill associated bacteria of two alvinocaridid species from different chemosynthetically ecological niches were first captured by single-molecule real-time sequencing. Totally, 120,792 optimized circular consensus sequences with ∼1,450 bp in length were obtained and clustered into 578 operational taxonomic units. Alpha diversity analysis showed seep A. longirostris had the highest species richness and evenness (average Chao1 = 213.68, Shannon = 3.39). Beta diversity analysis revealed that all samples were clearly divided into three groups, and microbial community of A. longirostris from seep and vent were more related than the other comparisons. By permutational multivariate analysis of variance, the most significant community compositional variance was detected between seep A. longirostris and vent S. leurokolos (R 2 = 0.731, P = 0.001). The taxon tags were further classified into 21 phyla, 40 classes, 89 orders, 124 families and 135 genera. Overall, the microbial communities were dominated by Campylobacteria and Gammaproteobacteria. Alphaproteobacteria, Bacteroidia, Verrucomicrobiae, Bacilli and other minor groups were also detected at lower abundance. Taxonomic groups recovered from the vent S. leurokolos samples were only dominated by Sulfurovaceae (94.06%). In comparison, gill-associated microbiota of vent A. longirostris consisted of more diverse sulfur-oxidizing bacteria, including Sulfurovaceae (69.21%), Thiotrichaceae (6.77%) and a putative novel Gammaproteobacteria group (14.37%), while in seep A. longirostris, Gammaproteobacteria un-group (44.01%) constituted the major component, following the methane-oxidizing bacteria Methylomonadaceae (19.38%), and Sulfurovaceae (18.66%). Therefore, the gill associated bacteria composition and abundance of alvinocaridid shrimps are closely related to the habitat heterogeneity and the selection of microbiota by the host. However, the interaction between these alvinocaridid shrimps and the epibiotic communities requires further study based on metagenome sequencing and fluorescence in situ hybridization.
Keywords: Alvinocarididae; Deep-sea; Hydrothermal vent; Methane seep; Microbial diversity.
©2022 Hui et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures




Similar articles
-
The complete mitochondrial genome of the alvinocaridid shrimp Shinkaicaris leurokolos (Decapoda, Caridea): Insight into the mitochondrial genetic basis of deep-sea hydrothermal vent adaptation in the shrimp.Comp Biochem Physiol Part D Genomics Proteomics. 2018 Mar;25:42-52. doi: 10.1016/j.cbd.2017.11.002. Epub 2017 Nov 10. Comp Biochem Physiol Part D Genomics Proteomics. 2018. PMID: 29145028
-
Insights into the strategy of micro-environmental adaptation: Transcriptomic analysis of two alvinocaridid shrimps at a hydrothermal vent.PLoS One. 2020 Jan 10;15(1):e0227587. doi: 10.1371/journal.pone.0227587. eCollection 2020. PLoS One. 2020. PMID: 31923275 Free PMC article.
-
First Comparative Analysis of the Community Structures and Carbon Metabolic Pathways of the Bacteria Associated with Alvinocaris longirostris in a Hydrothermal Vent of Okinawa Trough.PLoS One. 2016 Apr 25;11(4):e0154359. doi: 10.1371/journal.pone.0154359. eCollection 2016. PLoS One. 2016. PMID: 27111851 Free PMC article.
-
Complete mitochondrial genome of the hydrothermal vent shrimp Rimicaris variabilis (Decapoda: Caridea: Alvinocarididae) from the North Fiji basin.Mitochondrial DNA B Resour. 2019 Oct 11;4(2):3475-3476. doi: 10.1080/23802359.2019.1674717. Mitochondrial DNA B Resour. 2019. PMID: 33366046 Free PMC article.
-
Characterization of tissue-associated bacterial community of two Bathymodiolus species from the adjacent cold seep and hydrothermal vent environments.Sci Total Environ. 2021 Nov 20;796:149046. doi: 10.1016/j.scitotenv.2021.149046. Epub 2021 Jul 15. Sci Total Environ. 2021. PMID: 34328889
Cited by
-
Presence of an ultra-small microbiome in fermented cabbages.PeerJ. 2023 Jul 17;11:e15680. doi: 10.7717/peerj.15680. eCollection 2023. PeerJ. 2023. PMID: 37483986 Free PMC article.
-
Variation in epibiotic bacteria on two squat lobster species of Munidopsidae.Front Microbiol. 2023 Jun 28;14:1197476. doi: 10.3389/fmicb.2023.1197476. eCollection 2023. Front Microbiol. 2023. PMID: 37448572 Free PMC article.
-
Oral Microbial Dysbiosis Driven by Periodontitis Facilitates Oral Squamous Cell Carcinoma Progression.Cancers (Basel). 2025 Jun 28;17(13):2181. doi: 10.3390/cancers17132181. Cancers (Basel). 2025. PMID: 40647479 Free PMC article.
References
-
- Abràmoff MD, Magalhães PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11(7):36–42.
-
- Alain K, Olagnon M, Desbruyères D, Pagé A, Barbier G, Juniper SK, Quérellou J, Cambon-Bonavita MA. Phylogenetic characterization of the bacterial assemblage associated with mucous secretions of the hydrothermal vent polychaete Paralvinella palmiformis. FEMS Microbiology Ecology. 2002;42:463–476. doi: 10.1111/j.1574-6941.2002.tb01035.x. - DOI - PubMed
-
- Alayse-Danet AM, Desbruyeres D, Gaill F. The possible nutritional or detoxification role of the epibiotic bacteria of Alvinellid polychaetes: review of current data. Symbiosis. 1987;4:51–62.
-
- Apremont V, Cambon-Bonavita MA, Cueff-Gauchard V, François D, Pradillon F, Corbari L, Zbinden M. Gill chamber and gut microbial communities of the hydrothermal shrimp Rimicaris chacei Williams and Rona 1986: a possible symbiosis. PLOS ONE. 2018;13(11):e0206084. doi: 10.1371/journal.pone.0206084. - DOI - PMC - PubMed
-
- Beaulieu SE, Baker ET, German CR, Maffei A. An authoritative global database for active submarine hydrothermal vent fields. Geochemistry, Geophysics, Geosystems. 2013;14:11. doi: 10.1002/2013GC004998. - DOI
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources

