Dysbiotic human oral microbiota alters systemic metabolism via modulation of gut microbiota in germ-free mice
- PMID: 35966937
- PMCID: PMC9373767
- DOI: 10.1080/20002297.2022.2110194
Dysbiotic human oral microbiota alters systemic metabolism via modulation of gut microbiota in germ-free mice
Abstract
Background: The effect of oral microbiota on the intestinal microbiota has garnered growing attention as a mechanism linking periodontal diseases to systemic diseases. However, the salivary microbiota is diverse and comprises numerous bacteria with a largely similar composition in healthy individuals and periodontitis patients.
Aim: We explored how health-associated and periodontitis-associated salivary microbiota differently colonized the intestine and their subsequent systemic effects.
Methods: The salivary microbiota was collected from a healthy individual and a periodontitis patient and gavaged into C57BL/6NJcl[GF] mice. Gut microbial communities, hepatic gene expression profiles, and serum metabolites were analyzed.
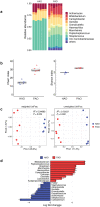
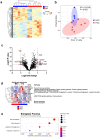
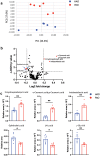
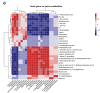
Results: The gut microbial composition was significantly different between periodontitis-associated microbiota-administered (PAO) and health-associated oral microbiota-administered (HAO) mice. The hepatic gene expression profile demonstrated a distinct pattern between the two groups, with higher expression of lipid and glucose metabolism-related genes. Disease-associated metabolites such as 2-hydroxyisobutyric acid and hydroxybenzoic acid were elevated in PAO mice. These metabolites were significantly correlated with characteristic gut microbial taxa in PAO mice. Conversely, health-associated oral microbiota were associated with higher levels of beneficial serum metabolites in HAO mice.
Conclusion: The multi-omics approach used in this study revealed that periodontitis-associated oral microbiota is associated with the induction of disease phenotype when they colonized the gut of germ-free mice.
Keywords: Oral; gut; liver; metabolome; microbiome; transcriptome.
© 2022 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
-
- Qin N, Yang F, Li A, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513(7516):59–14. Epub 2014/08/01. PubMed PMID: 25079328 - PubMed
-
- Read E, Curtis MA, Neves JF.. The role of oral bacteria in inflammatory bowel disease.Nat Rev Gastroenterol Hepatol.2021;18(10):731–742;Epub 2021/08/18. PubMed PMID: 34400822; - PubMed
-
- Yachida S, Mizutani S, Shiroma H, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25(6):968–976. Epub 2019/06/07. PubMed PMID: 31171880 - PubMed
-
- Kageyama S, Takeshita T, Asakawa M, et al. Relative abundance of total subgingival plaque-specific bacteria in salivary microbiota reflects the overall periodontal condition in patients with periodontitis. PLoS One. 2017;12(4):e0174782. Epub 2017/04/04. PubMed PMID: 28369125; PubMed Central PMCID: PMCPMC5378373 - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
