Single-cell RNA-Seq reveals changes in immune landscape in post-traumatic osteoarthritis
- PMID: 35967299
- PMCID: PMC9373730
- DOI: 10.3389/fimmu.2022.938075
Single-cell RNA-Seq reveals changes in immune landscape in post-traumatic osteoarthritis
Abstract
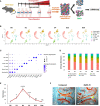
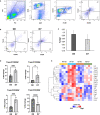
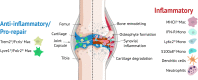
Osteoarthritis (OA) is the most common joint disease, affecting over 300 million people world-wide. Accumulating evidence attests to the important roles of the immune system in OA pathogenesis. Understanding the role of various immune cells in joint degeneration or joint repair after injury is vital for improving therapeutic strategies for treating OA. Post-traumatic osteoarthritis (PTOA) develops in ~50% of individuals who have experienced an articular trauma like an anterior cruciate ligament (ACL) rupture. Here, using the high resolution of single-cell RNA sequencing, we delineated the temporal dynamics of immune cell accumulation in the mouse knee joint after ACL rupture. Our study identified multiple immune cell types in the joint including neutrophils, monocytes, macrophages, B cells, T cells, NK cells and dendritic cells. Monocytes and macrophage populations showed the most dramatic changes after injury. Further characterization of monocytes and macrophages reveled 9 major subtypes with unique transcriptomics signatures, including a tissue resident Lyve1hiFolr2hi macrophage population and Trem2hiFcrls+ recruited macrophages, both showing enrichment for phagocytic genes and growth factors such as Igf1, Pdgfa and Pdgfc. We also identified several genes induced or repressed after ACL injury in a cell type-specific manner. This study provides new insight into PTOA-associated changes in the immune microenvironment and highlights macrophage subtypes that may play a role in joint repair after injury.
Keywords: Trem2; immune cells; inflammation; knee injury; macrophages; osteoarthritis; ptoa; scRNA-seq.
Copyright © 2022 Sebastian, Hum, McCool, Wilson, Murugesh, Martin, Rios-Arce, Amiri, Christiansen and Loots.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

