A Cross-Tissue Transcriptome-Wide Association Study Identifies Novel Susceptibility Genes for Juvenile Idiopathic Arthritis in Asia and Europe
- PMID: 35967305
- PMCID: PMC9367689
- DOI: 10.3389/fimmu.2022.941398
A Cross-Tissue Transcriptome-Wide Association Study Identifies Novel Susceptibility Genes for Juvenile Idiopathic Arthritis in Asia and Europe
Abstract
Background: Juvenile idiopathic arthritis (JIA) is the most common rheumatic disease in children, and its pathogenesis is still unclear. Genome-wide association studies (GWASs) of JIA have identified hundreds of risk factors, but few of them implicated specific biological mechanisms.
Methods: A cross-tissue transcriptome-wide association study (TWAS) was performed with the functional summary-based imputation software (FUSION) tool based on GWAS summary datasets (898 JIA patients and 346,102 controls from BioBank Japan (BBJ)/FinnGen). The gene expression reference weights of skeletal muscle and the whole blood were obtained from the Genotype-Tissue Expression (GTExv8) project. JIA-related genes identified by TWAS findings genes were further compared with the differentially expressed genes (DEGs) identified by the mRNA expression profile of JIA from the Gene Expression Omnibus (GEO) database (accession number: GSE1402). Last, candidate genes were analyzed using functional enrichment and annotation analysis by Metascape to examine JIA-related gene sets.
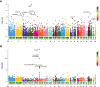
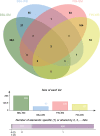
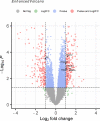
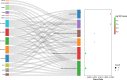
Results: The TWAS identified 535 significant genes with P < 0.05 and contains 350 for Asian and 195 for European (including 10 genes both expressed in Asian and European), such as CDC16 (P = 1.72E-03) and PSMD5-AS1 (P = 3.65E-02). Eight overlapping genes were identified based on TWAS results and DEGs of JIA patients, such as SIRPB1 (PTWAS = 4.21E-03, PDEG = 1.50E-04) and FRAT2 (PTWAS = 2.82E-02, PDEG = 1.43E-02). Pathway enrichment analysis of TWAS identified 183 pathways such as cytokine signaling in the immune system and cell adhesion molecules. By integrating the results of DEGs pathway and process enrichment analyses, 19 terms were identified such as positive regulation of T-cell activation.
Conclusion: By conducting two populations TWAS, we identified a group of JIA-associated genes and pathways, which may provide novel clues to uncover the pathogenesis of JIA.
Keywords: gene ontology; juvenile idiopathic arthritis; mRNA expression profiles; pathway enrichment; transcriptome-wide association study.
Copyright © 2022 Xu, Ma, Zeng, Si, Wu, Zhang and Shen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Integrative Analysis of Transcriptome-Wide Association Study and Gene-Based Association Analysis Identifies In Silico Candidate Genes Associated with Juvenile Idiopathic Arthritis.Int J Mol Sci. 2022 Nov 4;23(21):13555. doi: 10.3390/ijms232113555. Int J Mol Sci. 2022. PMID: 36362342 Free PMC article.
-
Identification of candidate genes and pathways associated with juvenile idiopathic arthritis by integrative transcriptome-wide association studies and mRNA expression profiles.Arthritis Res Ther. 2023 Feb 8;25(1):19. doi: 10.1186/s13075-023-03003-z. Arthritis Res Ther. 2023. PMID: 36755318 Free PMC article.
-
Integrating transcriptome-wide association study and mRNA expression profile identified candidate genes related to hand osteoarthritis.Arthritis Res Ther. 2021 Mar 10;23(1):81. doi: 10.1186/s13075-021-02458-2. Arthritis Res Ther. 2021. PMID: 33691763 Free PMC article.
-
The Multi-Omics Architecture of Juvenile Idiopathic Arthritis.Cells. 2020 Oct 15;9(10):2301. doi: 10.3390/cells9102301. Cells. 2020. PMID: 33076506 Free PMC article. Review.
-
Genetics of juvenile idiopathic arthritis: an update.Curr Opin Rheumatol. 2004 Sep;16(5):588-94. doi: 10.1097/01.bor.0000134407.48586.b0. Curr Opin Rheumatol. 2004. PMID: 15314499 Review.
Cited by
-
Large-scale integrative analysis of juvenile idiopathic arthritis for new insight into its pathogenesis.Arthritis Res Ther. 2024 Feb 10;26(1):47. doi: 10.1186/s13075-024-03280-2. Arthritis Res Ther. 2024. PMID: 38336809 Free PMC article.
-
Integrative Analysis of Transcriptome-Wide Association Study and Gene-Based Association Analysis Identifies In Silico Candidate Genes Associated with Juvenile Idiopathic Arthritis.Int J Mol Sci. 2022 Nov 4;23(21):13555. doi: 10.3390/ijms232113555. Int J Mol Sci. 2022. PMID: 36362342 Free PMC article.
-
The clinical and experimental treatment of Juvenile Idiopathic Arthritis.Clin Exp Immunol. 2023 Oct 13;213(3):276-287. doi: 10.1093/cei/uxad045. Clin Exp Immunol. 2023. PMID: 37074076 Free PMC article.
-
Integrative bioinformatics approach identifies novel drug targets for hyperaldosteronism, with a focus on SHMT1 as a promising therapeutic candidate.Sci Rep. 2025 Jan 11;15(1):1690. doi: 10.1038/s41598-025-85900-8. Sci Rep. 2025. PMID: 39799159 Free PMC article.
References
-
- Symmons DP, Jones M, Osborne J, Sills J, Southwood TR, Woo P. Pediatric Rheumatology in the United Kingdom: Data From the British Pediatric Rheumatology Group National Diagnostic Register. J Rheumatol (1996) 23:1975–80. - PubMed
-
- Petty RE, Southwood TR, Manners P, Baum J, Glass DN, Goldenberg J, et al. . International League of Associations for Rheumatology Classification of Juvenile Idiopathic Arthritis: Second Revision, Edmonton, 2001. J Rheumatol (2004) 31:390–2. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

