Peptide Centric Vβ Specific Germline Contacts Shape a Specialist T Cell Response
- PMID: 35967379
- PMCID: PMC9372435
- DOI: 10.3389/fimmu.2022.847092
Peptide Centric Vβ Specific Germline Contacts Shape a Specialist T Cell Response
Abstract
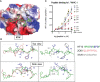
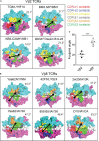
Certain CD8 T cell responses are particularly effective at controlling infection, as exemplified by elite control of HIV in individuals harboring HLA-B57. To understand the structural features that contribute to CD8 T cell elite control, we focused on a strongly protective CD8 T cell response directed against a parasite-derived peptide (HF10) presented by an atypical MHC-I molecule, H-2Ld. This response exhibits a focused TCR repertoire dominated by Vβ2, and a representative TCR (TG6) in complex with Ld-HF10 reveals an unusual structure in which both MHC and TCR contribute extensively to peptide specificity, along with a parallel footprint of TCR on its pMHC ligand. The parallel footprint is a common feature of Vβ2-containing TCRs and correlates with an unusual Vα-Vβ interface, CDR loop conformations, and Vβ2-specific germline contacts with peptides. Vβ2 and Ld may represent "specialist" components for antigen recognition that allows for particularly strong and focused T cell responses.
Keywords: MHC; TCR; elite controller; germline contacts; structure.
Copyright © 2022 Wang, Tsitsiklis, Devoe, Gao, Chu, Zhang, Li, Wong, Deane, Neau, Slansky, Thomas, Robey and Dai.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

