Seed Longevity in Legumes: Deeper Insights Into Mechanisms and Molecular Perspectives
- PMID: 35968115
- PMCID: PMC9364935
- DOI: 10.3389/fpls.2022.918206
Seed Longevity in Legumes: Deeper Insights Into Mechanisms and Molecular Perspectives
Abstract
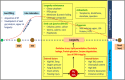
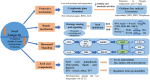
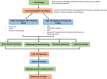
Sustainable agricultural production largely depends upon the viability and longevity of high-quality seeds during storage. Legumes are considered as rich source of dietary protein that helps to ensure nutritional security, but associated with poor seed longevity that hinders their performance and productivity in farmer's fields. Seed longevity is the key determinant to assure proper seed plant value and crop yield. Thus, maintenance of seed longevity during storage is of prime concern and a pre-requisite for enhancing crop productivity of legumes. Seed longevity is significantly correlated with other seed quality parameters such as germination, vigor, viability and seed coat permeability that affect crop growth and development, consequently distressing crop yield. Therefore, information on genetic basis and regulatory networks associated with seed longevity, as well as molecular dissection of traits linked to longevity could help in developing crop varieties with good storability. Keeping this in view, the present review focuses towards highlighting the molecular basis of seed longevity, with special emphasis on candidate genes and proteins associated with seed longevity and their interplay with other quality parameters. Further, an attempt was made to provide information on 3D structures of various genetic loci (genes/proteins) associated to seed longevity that could facilitate in understanding the interactions taking place within the seed at molecular level. This review compiles and provides information on genetic and genomic approaches for the identification of molecular pathways and key players involved in the maintenance of seed longevity in legumes, in a holistic manner. Finally, a hypothetical fast-forward breeding pipeline has been provided, that could assist the breeders to successfully develop varieties with improved seed longevity in legumes.
Keywords: crop productivity; genes; legumes; molecular pathways; seed longevity.
Copyright © 2022 Ramtekey, Cherukuri, Kumar, V., Sheoran, K., K., Kumar, Singh and Singh.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
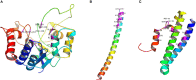
References
-
- Abdalla F. H., Roberts E. H. (1968). Effects of temperature, moisture, and oxygen on the induction of chromosome damage in seeds of barley, broad beans, and peas during storage. Ann. Bot. 32, 119–136. 10.1093/oxfordjournals.aob.a084187 - DOI
-
- Amara I., Zaidi I., Masmoudi K., Ludevid M. D., Pagès M., Goday A., et al. (2014). Insights into Late Embryogenesis Abundant (LEA) proteins in plants: from structure to the functions. Am. J. Plant Sci. 5, 3440–3455. 10.4236/ajps.2014.522360 - DOI
Publication types
LinkOut - more resources
Full Text Sources

