Current Advances in Paper-Based Biosensor Technologies for Rapid COVID-19 Diagnosis
- PMID: 35968255
- PMCID: PMC9363872
- DOI: 10.1007/s13206-022-00078-9
Current Advances in Paper-Based Biosensor Technologies for Rapid COVID-19 Diagnosis
Abstract
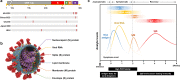
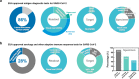
The global coronavirus disease 2019 (COVID-19) pandemic has had significant economic and social impacts on billions of people worldwide since severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first reported in Wuhan, China, in November 2019. Although polymerase chain reaction (PCR)-based technology serves as a robust test to detect SARS-CoV-2 in patients with COVID-19, there is a high demand for cost-effective, rapid, comfortable, and accurate point-of-care diagnostic tests in medical facilities. This review introduces the SARS-CoV-2 viral structure and diagnostic biomarkers derived from viral components. A comprehensive introduction of a paper-based diagnostic platform, including detection mechanisms for various target biomarkers and a COVID-19 commercial kit is presented. Intrinsic limitations related to the poor performance of currently developed paper-based devices and unresolved issues are discussed. Furthermore, we provide insight into novel paper-based diagnostic platforms integrated with advanced technologies such as nanotechnology, aptamers, surface-enhanced Raman spectroscopy (SERS), and clustered regularly interspaced short palindromic repeats (CRISPR)-Cas. Finally, we discuss the prospects for the development of highly sensitive, accurate, cost-effective, and easy-to-use point-of-care COVID-19 diagnostic methods.
Keywords: COVID-19; Lateral flow assay (LFA); Paper-based biosensors; Point-of-care testing (POCT); SARS-CoV-2.
© The Korean BioChip Society 2022, Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Conflict of interest statement
Conflict of interestThe authors declare that they have no conflict of interest.
Figures




Similar articles
-
Recent advances in sensitive surface-enhanced Raman scattering-based lateral flow assay platforms for point-of-care diagnostics of infectious diseases.Sens Actuators B Chem. 2021 Feb 15;329:129214. doi: 10.1016/j.snb.2020.129214. Epub 2020 Nov 19. Sens Actuators B Chem. 2021. PMID: 36568647 Free PMC article.
-
[Recent advances in clustered regularly interspaced short palindromic repeats-based detection of severe acute respiratory syndrome coronavirus 2].Se Pu. 2022 Sep;40(9):773-781. doi: 10.3724/SP.J.1123.2022.08001. Se Pu. 2022. PMID: 36156623 Free PMC article. Review. Chinese.
-
Sensitive and rapid on-site detection of SARS-CoV-2 using a gold nanoparticle-based high-throughput platform coupled with CRISPR/Cas12-assisted RT-LAMP.Sens Actuators B Chem. 2021 Oct 15;345:130411. doi: 10.1016/j.snb.2021.130411. Epub 2021 Jul 6. Sens Actuators B Chem. 2021. PMID: 34248284 Free PMC article.
-
Advancements in Detection Approaches of Severe Acute Respiratory Syndrome Coronavirus 2.Malays J Med Sci. 2022 Dec;29(6):15-33. doi: 10.21315/mjms2022.29.6.3. Epub 2022 Dec 22. Malays J Med Sci. 2022. PMID: 36818907 Free PMC article. Review.
-
CLEVER assay: A visual and rapid RNA extraction-free detection of SARS-CoV-2 based on CRISPR-Cas integrated RT-LAMP technology.J Appl Microbiol. 2022 Aug;133(2):410-421. doi: 10.1111/jam.15571. Epub 2022 Apr 18. J Appl Microbiol. 2022. PMID: 35396760 Free PMC article.
Cited by
-
Aptamer-Based Point-of-Care Devices: Emerging Technologies and Integration of Computational Methods.Biosensors (Basel). 2023 May 22;13(5):569. doi: 10.3390/bios13050569. Biosensors (Basel). 2023. PMID: 37232930 Free PMC article. Review.
-
Preventing SARS-CoV-2 infection using Fv-antibodies targeting the proprotein convertase (PPC) cleavage site.RSC Med Chem. 2024 Aug 26;15(11):3704-10. doi: 10.1039/d4md00552j. Online ahead of print. RSC Med Chem. 2024. PMID: 39290379 Free PMC article.
-
Lipid-based colloidal nanoparticles for applications in targeted vaccine delivery.Nanoscale Adv. 2023 Feb 17;5(7):1853-1869. doi: 10.1039/d2na00795a. eCollection 2023 Mar 28. Nanoscale Adv. 2023. PMID: 36998671 Free PMC article. Review.
-
Application of microfluidic technologies on COVID-19 diagnosis and drug discovery.Acta Pharm Sin B. 2023 Feb 24;13(7):2877-96. doi: 10.1016/j.apsb.2023.02.014. Online ahead of print. Acta Pharm Sin B. 2023. PMID: 36855672 Free PMC article. Review.
-
Updates on the Biofunctionalization of Gold Nanoparticles for the Rapid and Sensitive Multiplatform Diagnosis of SARS-CoV-2 Virus and Its Proteins: From Computational Models to Validation in Human Samples.Int J Mol Sci. 2023 May 25;24(11):9249. doi: 10.3390/ijms24119249. Int J Mol Sci. 2023. PMID: 37298201 Free PMC article. Review.
References
-
- Johns Hopkins University (JHU) and Center for Systems Science and Engineering (CSSE), Coronavirus Resource Center., cited April 28, 2022. https://coronavirus.jhu.edu/map.html
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
