An Adjuvant-Based Approach Enables the Use of Dominant HYG and KAN Selectable Markers in Candida albicans
- PMID: 35968963
- PMCID: PMC9429937
- DOI: 10.1128/msphere.00347-22
An Adjuvant-Based Approach Enables the Use of Dominant HYG and KAN Selectable Markers in Candida albicans
Abstract
Candida albicans is a pathobiont fungus that can colonize multiple niches in the human body but is also a frequent cause of both mucosal and systemic disease. Despite its clinical importance, a paucity of dominant selectable markers has hindered the development of tools for genetic manipulation of the species. One factor limiting the utilization of dominant selectable markers is that C. albicans is inherently more resistant to antibiotics used for selection in other species. Here, we showed that the inclusion of suitable adjuvants can enable the use of two aminoglycoside antibiotics, hygromycin B and G418, for positive selection in C. albicans. Combining these antibiotics with an adjuvant, such as quinine or molybdate, substantially suppressed the background growth of C. albicans, thereby enabling transformants expressing CaHygB or CaKan markers to be readily identified. We verified that these adjuvants were not mutagenic to C. albicans and that CaHygB and CaKan markers were orthogonal to the existing marker NAT1/SAT1, and so provide complementary tools for the genetic manipulation of C. albicans strains. Our study also established that adjuvant-based approaches can enable the use of selectable markers that would otherwise be limited by high background growth from susceptible cells. IMPORTANCE Only a single dominant selectable marker has been widely adopted for use in the opportunistic fungal pathogen Candida albicans. This is in stark contrast to model fungi where a repertoire of dominant markers is readily available. A limiting factor for C. albicans has been the high levels of background growth obtained with multiple antibiotics, thereby limiting their use for distinguishing cells that carry an antibiotic-resistance gene from those that do not. Here, we demonstrated that the inclusion of adjuvants can reduce background growth and enable the robust use of both CaHygB and CaKan markers for genetic selection in C. albicans.
Keywords: antibiotic selection; dominant marker; drug resistance; fungal pathogen; selectable marker.
Conflict of interest statement
The authors declare a conflict of interest. A provisional patent on the use of adjuvants to enable the use of antibiotic selectable markers has been submitted by the authors.
Figures

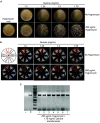



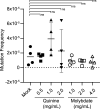
References
-
- Kett DH, Azoulay E, Echeverria PM, Vincent JL, Extended Prevalence of Infection in ICUSGoI. 2011. Candida bloodstream infections in intensive care units: analysis of the extended prevalence of infection in intensive care unit study. Crit Acre Med 39:665–670. doi:10.1097/CCM.0b013e318206c1ca. - DOI - PubMed
-
- Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, Agrafioti I, Arnaud MB, Bates S, Brown AJ, Brunke S, Costanzo MC, Fitzpatrick DA, de Groot PW, Harris D, Hoyer LL, Hube B, Klis FM, Kodira C, Lennard N, Logue ME, Martin R, Neiman AM, Nikolaou E, Quail MA, Quinn J, Santos MC, Schmitzberger FF, Sherlock G, Shah P, Silverstein KA, Skrzypek MS, Soll D, Staggs R, Stansfield I, Stumpf MP, Sudbery PE, Srikantha T, Zeng Q, Berman J, Berriman M, Heitman J, Gow NA, Lorenz MC, Birren BW, Kellis M, et al. . 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459:657–662. doi:10.1038/nature08064. - DOI - PMC - PubMed
-
- Krassowski T, Coughlan AY, Shen XX, Zhou X, Kominek J, Opulente DA, Riley R, Grigoriev IV, Maheshwari N, Shields DC, Kurtzman CP, Hittinger CT, Rokas A, Wolfe KH. 2018. Evolutionary instability of CUG-Leu in the genetic code of budding yeasts. Nat Commun 9:1887. doi:10.1038/s41467-018-04374-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
