Tissue-specific modifier alleles determine Mertk loss-of-function traits
- PMID: 35969037
- PMCID: PMC9433089
- DOI: 10.7554/eLife.80530
Tissue-specific modifier alleles determine Mertk loss-of-function traits
Abstract
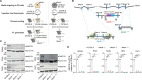
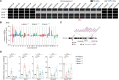
Knockout (KO) mouse models play critical roles in elucidating biological processes behind disease-associated or disease-resistant traits. As a presumed consequence of gene KO, mice display certain phenotypes. Based on insight into the molecular role of said gene in a biological process, it is inferred that the particular biological process causally underlies the trait. This approach has been crucial towards understanding the basis of pathological and/or advantageous traits associated with Mertk KO mice. Mertk KO mice suffer from severe, early-onset retinal degeneration. MERTK, expressed in retinal pigment epithelia, is a receptor tyrosine kinase with a critical role in phagocytosis of apoptotic cells or cellular debris. Therefore, early-onset, severe retinal degeneration was described to be a direct consequence of failed MERTK-mediated phagocytosis of photoreceptor outer segments by retinal pigment epithelia. Here, we report that the loss of Mertk alone is not sufficient for retinal degeneration. The widely used Mertk KO mouse carries multiple coincidental changes in its genome that affect the expression of a number of genes, including the Mertk paralog Tyro3. Retinal degeneration manifests only when the function of Tyro3 is concomitantly lost. Furthermore, Mertk KO mice display improved anti-tumor immunity. MERTK is expressed in macrophages. Therefore, enhanced anti-tumor immunity was inferred to result from the failure of macrophages to dispose of cancer cell corpses, resulting in a pro-inflammatory tumor microenvironment. The resistance against two syngeneic mouse tumor models observed in Mertk KO mice is not, however, phenocopied by the loss of Mertk alone. Neither Tyro3 nor macrophage phagocytosis by alternate genetic redundancy accounts for the absence of anti-tumor immunity. Collectively, our results indicate that context-dependent epistasis of independent modifier alleles determines Mertk KO traits.
Keywords: Mertk knockout mice; anti-tumor immunity; immunology; inflammation; mouse; retinal degeneration.
© 2022, Akalu, Mercau et al.
Conflict of interest statement
YA, MM, MA, LH, JN, EA, XL, YK, WP, MB, SF, AI, AL No competing interests declared, LH, DA, TK is affiliated with Celldex Therapeutics. The author has no financial interests to declare, CR Senior editor, eLife, SG has received grant support from Mirati Therapeutics
Figures








References
-
- Camenisch TD, Koller BH, Earp HS, Matsushima GK. A novel receptor tyrosine kinase, mer, inhibits TNF-alpha production and lipopolysaccharide-induced endotoxic shock. Journal of Immunology. 1999;162:3498–3503. - PubMed
-
- Chan PY, Carrera Silva EA, De Kouchkovsky D, Joannas LD, Hao L, Hu D, Huntsman S, Eng C, Licona-Limón P, Weinstein JS, Herbert DR, Craft JE, Flavell RA, Repetto S, Correale J, Burchard EG, Torgerson DG, Ghosh S, Rothlin CV. The TAM family receptor tyrosine kinase TYRO3 is a negative regulator of type 2 immunity. Science. 2016;352:99–103. doi: 10.1126/science.aaf1358. - DOI - PMC - PubMed
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

