Integrated 3d flow-based multi-atlas brain structure segmentation
- PMID: 35969596
- PMCID: PMC9377636
- DOI: 10.1371/journal.pone.0270339
Integrated 3d flow-based multi-atlas brain structure segmentation
Abstract
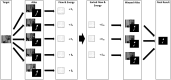
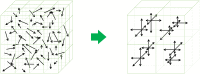
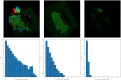
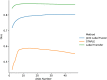
MRI brain structure segmentation plays an important role in neuroimaging studies. Existing methods either spend much CPU time, require considerable annotated data, or fail in segmenting volumes with large deformation. In this paper, we develop a novel multi-atlas-based algorithm for 3D MRI brain structure segmentation. It consists of three modules: registration, atlas selection and label fusion. Both registration and label fusion leverage an integrated flow based on grayscale and SIFT features. We introduce an effective and efficient strategy for atlas selection by employing the accompanying energy generated in the registration step. A 3D sequential belief propagation method and a 3D coarse-to-fine flow matching approach are developed in both registration and label fusion modules. The proposed method is evaluated on five public datasets. The results show that it has the best performance in almost all the settings compared to competitive methods such as ANTs, Elastix, Learning to Rank and Joint Label Fusion. Moreover, our registration method is more than 7 times as efficient as that of ANTs SyN, while our label transfer method is 18 times faster than Joint Label Fusion in CPU time. The results on the ADNI dataset demonstrate that our method is applicable to image pairs that require a significant transformation in registration. The performance on a composite dataset suggests that our method succeeds in a cross-modality manner. The results of this study show that the integrated 3D flow-based method is effective and efficient for brain structure segmentation. It also demonstrates the power of SIFT features, multi-atlas segmentation and classical machine learning algorithms for a medical image analysis task. The experimental results on public datasets show the proposed method's potential for general applicability in various brain structures and settings.
Conflict of interest statement
All authors declare that YX had financial support from the National Natural Science Foundation in China, and the State Key Laboratory of Software Development Environment in Beihang University in China; no financial relationships with any organizations that might have an interest in the submitted work in the previous three years; no other relationships or activities that could appear to have influenced the submitted work. All funding affiliations do not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures








References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

