Transcription-replication conflicts in primordial germ cells necessitate the Fanconi anemia pathway to safeguard genome stability
- PMID: 35969748
- PMCID: PMC9407672
- DOI: 10.1073/pnas.2203208119
Transcription-replication conflicts in primordial germ cells necessitate the Fanconi anemia pathway to safeguard genome stability
Abstract
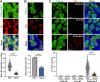
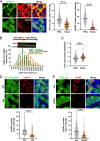
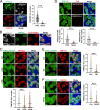
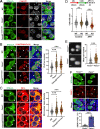
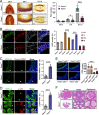
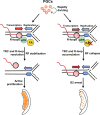
Preserving a high degree of genome integrity and stability in germ cells is of utmost importance for reproduction and species propagation. However, the regulatory mechanisms of maintaining genome stability in the developing primordial germ cells (PGCs), in which rapid proliferation is coupled with global hypertranscription, remain largely unknown. Here, we find that mouse PGCs encounter a constitutively high frequency of transcription-replication conflicts (TRCs), which lead to R-loop accumulation and impose endogenous replication stress on PGCs. We further demonstrate that the Fanconi anemia (FA) pathway is activated by TRCs and has a central role in the coordination between replication and transcription in the rapidly proliferating PGCs, as disabling the FA pathway leads to TRC and R-loop accumulation, replication fork destabilization, increased DNA damage, dramatic loss of mitotically dividing mouse PGCs, and consequent sterility of both sexes. Overall, our findings uncover the unique source and resolving mechanism of endogenous replication stress during PGC proliferation, provide a biological explanation for reproductive defects in individuals with FA, and improve our understanding of the monitoring strategies for genome stability during germ cell development.
Keywords: Fanconi anemia pathway; genome stability; primordial germ cells; replication stress; transcription–replication conflicts.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
-
- Suh E. K., et al. , p63 protects the female germ line during meiotic arrest. Nature 444, 624–628 (2006). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

