On the formation of ordered protein assemblies in cell-cell interfaces
- PMID: 35969779
- PMCID: PMC9407605
- DOI: 10.1073/pnas.2206175119
On the formation of ordered protein assemblies in cell-cell interfaces
Abstract
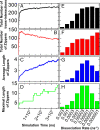
Crystal structures of many cell-cell adhesion receptors reveal the formation of linear "molecular zippers" comprising an ordered one-dimensional array of proteins that form both intercellular (trans) and intracellular (cis) interactions. The clustered protocadherins (cPcdhs) provide an exemplar of this phenomenon and use it as a basis of barcoding of vertebrate neurons. Here, we report both Metropolis and kinetic Monte Carlo simulations of cPcdh zipper formation using simplified models of cPcdhs that nevertheless capture essential features of their three-dimensional structure. The simulations reveal that the formation of long zippers is an implicit feature of cPcdh structure and is driven by their cis and trans interactions that have been quantitatively characterized in previous work. Moreover, in agreement with cryo-electron tomography studies, the zippers are found to organize into two-dimensional arrays even in the absence of attractive interactions between individual zippers. Our results suggest that the formation of ordered two-dimensional arrays of linear zippers of adhesion proteins is a common feature of cell-cell interfaces. From the perspective of simulations, they demonstrate the importance of a realistic depiction of adhesion protein structure and interactions if important biological phenomena are to be properly captured.
Keywords: adhesion proteins; cell–cell interfaces; clustered protocadherins; ordered protein assemblies.
Conflict of interest statement
The authors declare no competing interest.
Figures





References
-
- Grakoui A., et al. , The immunological synapse: A molecular machine controlling T cell activation. Science 285, 221–227 (1999). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

