ORP5 and ORP8 orchestrate lipid droplet biogenesis and maintenance at ER-mitochondria contact sites
- PMID: 35969857
- PMCID: PMC9375143
- DOI: 10.1083/jcb.202112107
ORP5 and ORP8 orchestrate lipid droplet biogenesis and maintenance at ER-mitochondria contact sites
Abstract
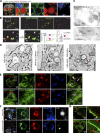
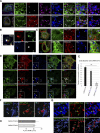
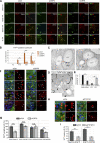
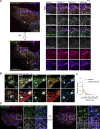
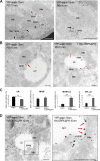
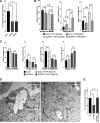
Lipid droplets (LDs) are the primary organelles of lipid storage, buffering energy fluctuations of the cell. They store neutral lipids in their core that is surrounded by a protein-decorated phospholipid monolayer. LDs arise from the endoplasmic reticulum (ER). The ER protein seipin, localizing at ER-LD junctions, controls LD nucleation and growth. However, how LD biogenesis is spatially and temporally coordinated remains elusive. Here, we show that the lipid transfer proteins ORP5 and ORP8 control LD biogenesis at mitochondria-associated ER membrane (MAM) subdomains, enriched in phosphatidic acid. We found that ORP5/8 regulates seipin recruitment to these MAM-LD contacts, and their loss impairs LD biogenesis. Importantly, the integrity of ER-mitochondria contact sites is crucial for ORP5/8 function in regulating seipin-mediated LD biogenesis. Our study uncovers an unprecedented ORP5/8 role in orchestrating LD biogenesis and maturation at MAMs and brings novel insights into the metabolic crosstalk between mitochondria, ER, and LDs at the membrane contact sites.
© 2022 Guyard et al.
Figures









Similar articles
-
ORP5 localizes to ER-lipid droplet contacts and regulates the level of PI(4)P on lipid droplets.J Cell Biol. 2020 Jan 6;219(1):e201905162. doi: 10.1083/jcb.201905162. J Cell Biol. 2020. PMID: 31653673 Free PMC article.
-
ORP5/ORP8 localize to endoplasmic reticulum-mitochondria contacts and are involved in mitochondrial function.EMBO Rep. 2016 Jun;17(6):800-10. doi: 10.15252/embr.201541108. Epub 2016 Apr 13. EMBO Rep. 2016. PMID: 27113756 Free PMC article.
-
Lipid Droplet Nucleation.Trends Cell Biol. 2021 Feb;31(2):108-118. doi: 10.1016/j.tcb.2020.11.006. Epub 2020 Dec 5. Trends Cell Biol. 2021. PMID: 33293168 Review.
-
Seipin collaborates with the ER membrane to control the sites of lipid droplet formation.Curr Opin Cell Biol. 2022 Apr;75:102070. doi: 10.1016/j.ceb.2022.02.004. Epub 2022 Mar 17. Curr Opin Cell Biol. 2022. PMID: 35306312 Free PMC article. Review.
-
Born this way - Biogenesis of lipid droplets from specialized ER subdomains.Biochim Biophys Acta Mol Cell Biol Lipids. 2020 Jan;1865(1):158448. doi: 10.1016/j.bbalip.2019.04.008. Epub 2019 Apr 24. Biochim Biophys Acta Mol Cell Biol Lipids. 2020. PMID: 31028912 Review.
Cited by
-
Transmembrane Membrane Readers form a Novel Class of Proteins That Include Peripheral Phosphoinositide Recognition Domains and Viral Spikes.Membranes (Basel). 2022 Nov 17;12(11):1161. doi: 10.3390/membranes12111161. Membranes (Basel). 2022. PMID: 36422153 Free PMC article. Review.
-
Cool-contacts: Cryo-Electron Microscopy of Membrane Contact Sites and Their Components.Contact (Thousand Oaks). 2024 Feb 25;7:25152564241231364. doi: 10.1177/25152564241231364. eCollection 2024 Jan-Dec. Contact (Thousand Oaks). 2024. PMID: 38410695 Free PMC article. Review.
-
Cell lipid biology in infections: an overview.Front Cell Infect Microbiol. 2023 Oct 6;13:1148383. doi: 10.3389/fcimb.2023.1148383. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37868347 Free PMC article. Review.
-
Current research on mitochondria‑associated membranes in cardiovascular diseases (Review).Mol Med Rep. 2025 Jun;31(6):141. doi: 10.3892/mmr.2025.13506. Epub 2025 Apr 4. Mol Med Rep. 2025. PMID: 40183396 Free PMC article. Review.
-
Ultrastructure analysis of mitochondria, lipid droplet and sarcoplasmic reticulum apposition in human heart failure.J Mol Cell Cardiol Plus. 2025 Jun 10;13:100461. doi: 10.1016/j.jmccpl.2025.100461. eCollection 2025 Sep. J Mol Cell Cardiol Plus. 2025. PMID: 40584838 Free PMC article.
References
-
- Ajjaji, D., Ben M’barek K., Mimmack M.L., England C., Herscovitz H., Dong L., Kay R.G., Patel S., Saudek V., Small D.M., et al. . 2019. Dual binding motifs underpin the hierarchical association of perilipins1-3 with lipid droplets. Mol. Biol. Cell. 30:703–716. 10.1091/mbc.E18-08-0534 - DOI - PMC - PubMed
-
- Benador, I.Y., Veliova M., Mahdaviani K., Petcherski A., Wikstrom J.D., Assali E.A., Acín-Pérez R., Shum M., Oliveira M.F., Cinti S., et al. . 2018. Mitochondria bound to lipid droplets have unique bioenergetics, composition, and dynamics that support lipid droplet expansion. Cell Metab. 27:869–885.e6. 10.1016/j.cmet.2018.03.003 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

