Allele specific PCR for a major marker of levamisole resistance in Haemonchus contortus
- PMID: 35970104
- PMCID: PMC9399269
- DOI: 10.1016/j.ijpddr.2022.08.001
Allele specific PCR for a major marker of levamisole resistance in Haemonchus contortus
Abstract
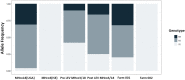
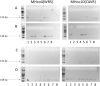
Haemonchus contortus is a haematophagous parasitic nematode that infects small ruminants and causes significant animal health concerns and economic losses within the livestock industry on a global scale. Treatment primarily depends on broad-spectrum anthelmintics, however, resistance is established or rapidly emerging against all major drug classes. Levamisole (LEV) remains an important treatment option for parasite control, as resistance to LEV is less prevalent than to members of other major classes of anthelmintics. LEV is an acetylcholine receptor (AChR) agonist that, when bound, results in paralysis of the worm. Numerous studies implicated the AChR sub-unit, ACR-8, in LEV sensitivity and in particular, the presence of a truncated acr-8 transcript or a deletion in the acr-8 locus in some resistant isolates. Recently, a single non-synonymous SNP in acr-8 conferring a serine-to-threonine substitution (S168T) was identified that was strongly associated with LEV resistance. Here, we investigate the role of genetic variation at the acr-8 locus in a controlled genetic cross between the LEV susceptible MHco3(ISE) and LEV resistant MHco18(UGA2004) isolates of H. contortus. Using single worm PCR assays, we found that the presence of S168T was strongly associated with LEV resistance in the parental isolates and F3 progeny of the genetic cross surviving LEV treatment. We developed and optimised an allele-specific PCR assay for the detection of S168T and validated the assay using laboratory isolates and field samples that were phenotyped for LEV resistance. In the LEV-resistant field population, a high proportion (>75%) of L3 encoded the S168T variant, whereas the variant was absent in the susceptible isolates studied. These data further support the potential role of acr-8 S168T in LEV resistance, with the allele-specific PCR providing an important step towards establishing a sensitive molecular diagnostic test for LEV resistance.
Keywords: Allele specific PCR; Diagnostic; Haemonchus; Levamisole; RFLP; Resistance; S168T; SNP.
Copyright © 2022 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors report that they have no conflict of interest.
Figures







References
-
- Avramenko R.W., Redman E.M., Melville L., Bartley Y., Wit J., Queiroz C., Bartley D.J., Gilleard J.S. Deep amplicon sequencing as a powerful new tool to screen for sequence polymorphisms associated with anthelmintic resistance in parasitic nematode populations. Int. J. Parasitol. 2019;49:13–26. - PubMed
-
- Barrère V., Beech R.N., Charvet C.L., Prichard R.K. Novel assay for the detection and monitoring of levamisole resistance in Haemonchus contortus. Int. J. Parasitol. 2014;44:235–241. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

