The Mis6 inner kinetochore subcomplex maintains CENP-A nucleosomes against centromeric non-coding transcription during mitosis
- PMID: 35970865
- PMCID: PMC9378642
- DOI: 10.1038/s42003-022-03786-y
The Mis6 inner kinetochore subcomplex maintains CENP-A nucleosomes against centromeric non-coding transcription during mitosis
Abstract
Centromeres are established by nucleosomes containing the histone H3 variant CENP-A. CENP-A is recruited to centromeres by the Mis18-HJURP machinery. During mitosis, CENP-A recruitment ceases, implying the necessity of CENP-A maintenance at centromeres, although the exact underlying mechanism remains elusive. Herein, we show that the inner kinetochore protein Mis6 (CENP-I) and Mis15 (CENP-N) retain CENP-A during mitosis in fission yeast. Eliminating Mis6 or Mis15 during mitosis caused immediate loss of pre-existing CENP-A at centromeres. CENP-A loss occurred due to the transcriptional upregulation of non-coding RNAs at the central core region of centromeres, as confirmed by the observation RNA polymerase II inhibition preventing CENP-A loss from centromeres in the mis6 mutant. Thus, we concluded that the inner kinetochore complex containing Mis6-Mis15 blocks the indiscriminate transcription of non-coding RNAs at the core centromere, thereby retaining the epigenetic inheritance of CENP-A during mitosis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


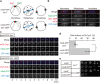

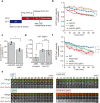
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

