Deep polygenic neural network for predicting and identifying yield-associated genes in Indonesian rice accessions
- PMID: 35970979
- PMCID: PMC9378700
- DOI: 10.1038/s41598-022-16075-9
Deep polygenic neural network for predicting and identifying yield-associated genes in Indonesian rice accessions
Abstract
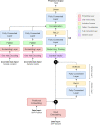
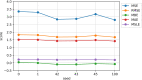
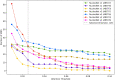
As the fourth most populous country in the world, Indonesia must increase the annual rice production rate to achieve national food security by 2050. One possible solution comes from the nanoscopic level: a genetic variant called Single Nucleotide Polymorphism (SNP), which can express significant yield-associated genes. The prior benchmark of this study utilized a statistical genetics model where no SNP position information and attention mechanism were involved. Hence, we developed a novel deep polygenic neural network, named the NucleoNet model, to address these obstacles. The NucleoNets were constructed with the combination of prominent components that include positional SNP encoding, the context vector, wide models, Elastic Net, and Shannon's entropy loss. This polygenic modeling obtained up to 2.779 of Mean Squared Error (MSE) with 47.156% of Symmetric Mean Absolute Percentage Error (SMAPE), while revealing 15 new important SNPs. Furthermore, the NucleoNets reduced the MSE score up to 32.28% compared to the Ordinary Least Squares (OLS) model. Through the ablation study, we learned that the combination of Xavier distribution for weights initialization and Normal distribution for biases initialization sparked more various important SNPs throughout 12 chromosomes. Our findings confirmed that the NucleoNet model was successfully outperformed the OLS model and identified important SNPs to Indonesian rice yields.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
RiceSNP-ABST: a deep learning approach to identify abiotic stress-associated single nucleotide polymorphisms in rice.Brief Bioinform. 2024 Nov 22;26(1):bbae702. doi: 10.1093/bib/bbae702. Brief Bioinform. 2024. PMID: 39757606 Free PMC article.
-
Genome-wide association mapping of salinity tolerance in rice (Oryza sativa).DNA Res. 2015 Apr;22(2):133-45. doi: 10.1093/dnares/dsu046. Epub 2015 Jan 27. DNA Res. 2015. PMID: 25627243 Free PMC article.
-
An improved 7K SNP array, the C7AIR, provides a wealth of validated SNP markers for rice breeding and genetics studies.PLoS One. 2020 May 14;15(5):e0232479. doi: 10.1371/journal.pone.0232479. eCollection 2020. PLoS One. 2020. PMID: 32407369 Free PMC article.
-
[Single nucleotide polymorphism (SNP) and its application in rice].Yi Chuan. 2006 Jun;28(6):737-44. Yi Chuan. 2006. PMID: 16818440 Review. Chinese.
-
Advances in genome-wide association studies of complex traits in rice.Theor Appl Genet. 2020 May;133(5):1415-1425. doi: 10.1007/s00122-019-03473-3. Epub 2019 Nov 12. Theor Appl Genet. 2020. PMID: 31720701 Review.
Cited by
-
Transformer Architecture and Attention Mechanisms in Genome Data Analysis: A Comprehensive Review.Biology (Basel). 2023 Jul 22;12(7):1033. doi: 10.3390/biology12071033. Biology (Basel). 2023. PMID: 37508462 Free PMC article. Review.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

