Sirt2 promotes white matter oligodendrogenesis during development and in models of neonatal hypoxia
- PMID: 35970992
- PMCID: PMC9378658
- DOI: 10.1038/s41467-022-32462-2
Sirt2 promotes white matter oligodendrogenesis during development and in models of neonatal hypoxia
Abstract
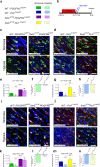
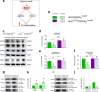
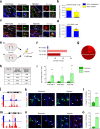
Delayed oligodendrocyte (OL) maturation caused by hypoxia (Hx)-induced neonatal brain injury results in hypomyelination and leads to neurological disabilities. Previously, we characterized Sirt1 as a crucial regulator of OL progenitor cell (OPC) proliferation in response to Hx. We now identify Sirt2 as a critical promoter of OL differentiation during both normal white matter development and in a mouse model of Hx. Importantly, we find that Hx reduces Sirt2 expression in mature OLs and that Sirt2 overexpression in OPCs restores mature OL populations. Reduced numbers of Sirt2+ OLs were also observed in the white matter of preterm human infants. We show that Sirt2 interacts with p27Kip1/FoxO1, p21Cip1/Cdk4, and Cdk5 pathways, and that these interactions are altered by Hx. Furthermore, Hx induces nuclear translocation of Sirt2 in OPCs where it binds several genomic targets. Overall, these results indicate that a balance of Sirt1 and Sirt2 activity is required for developmental oligodendrogenesis, and that these proteins represent potential targets for promoting repair following white matter injury.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

