Metagenomic features of bioburden serve as outcome indicators in combat extremity wounds
- PMID: 35970993
- PMCID: PMC9378645
- DOI: 10.1038/s41598-022-16170-x
Metagenomic features of bioburden serve as outcome indicators in combat extremity wounds
Abstract
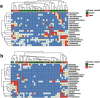
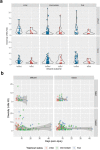
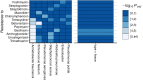
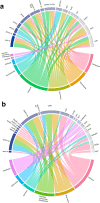
Battlefield injury management requires specialized care, and wound infection is a frequent complication. Challenges related to characterizing relevant pathogens further complicates treatment. Applying metagenomics to wounds offers a comprehensive path toward assessing microbial genomic fingerprints and could indicate prognostic variables for future decision support tools. Wound specimens from combat-injured U.S. service members, obtained during surgical debridements before delayed wound closure, were subjected to whole metagenome analysis and targeted enrichment of antimicrobial resistance genes. Results did not indicate a singular, common microbial metagenomic profile for wound failure, instead reflecting a complex microenvironment with varying bioburden diversity across outcomes. Genus-level Pseudomonas detection was associated with wound failure at all surgeries. A logistic regression model was fit to the presence and absence of antimicrobial resistance classes to assess associations with nosocomial pathogens. A. baumannii detection was associated with detection of genomic signatures for resistance to trimethoprim, aminoglycosides, bacitracin, and polymyxin. Machine learning classifiers were applied to identify wound and microbial variables associated with outcome. Feature importance rankings averaged across models indicated the variables with the largest effects on predicting wound outcome, including an increase in P. putida sequence reads. These results describe the microbial genomic determinants in combat wound bioburden and demonstrate metagenomic investigation as a comprehensive tool for providing information toward aiding treatment of combat-related injuries.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures