Antibiotics Drive Expansion of Rare Pathogens in a Chronic Infection Microbiome Model
- PMID: 35972133
- PMCID: PMC9599657
- DOI: 10.1128/msphere.00318-22
Antibiotics Drive Expansion of Rare Pathogens in a Chronic Infection Microbiome Model
Abstract
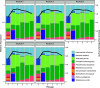
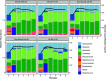
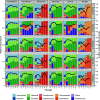
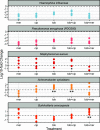
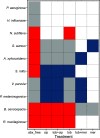
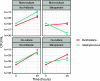
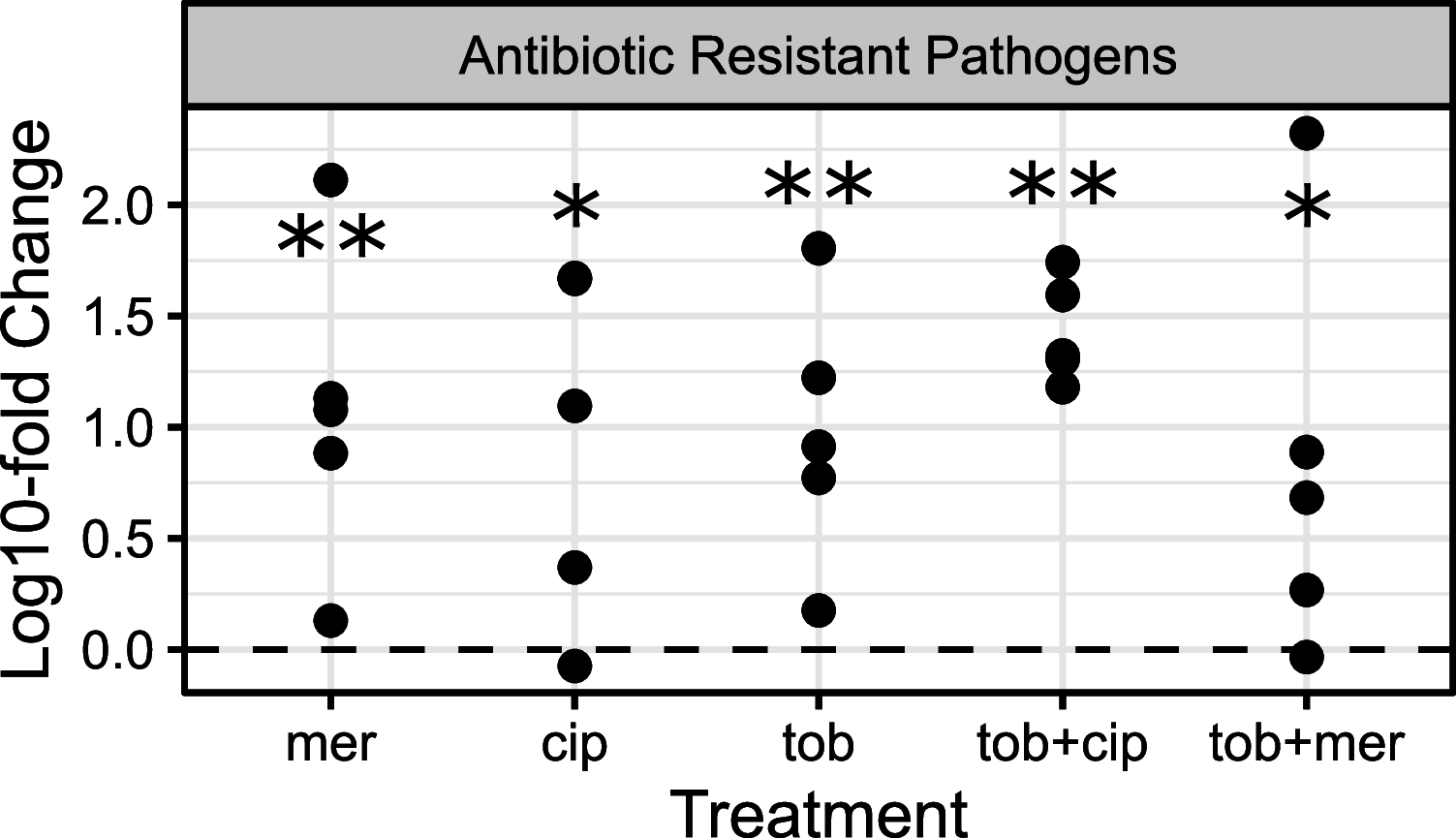
Chronic (long-lasting) infections are globally a major and rising cause of morbidity and mortality. Unlike typical acute infections, chronic infections are ecologically diverse, characterized by the presence of a polymicrobial mix of opportunistic pathogens and human-associated commensals. To address the challenge of chronic infection microbiomes, we focus on a particularly well-characterized disease, cystic fibrosis (CF), where polymicrobial lung infections persist for decades despite frequent exposure to antibiotics. Epidemiological analyses point to conflicting results on the benefits of antibiotic treatment yet are confounded by the dependency of antibiotic exposures on prior pathogen presence, limiting their ability to draw causal inferences on the relationships between antibiotic exposure and pathogen dynamics. To address this limitation, we develop a synthetic infection microbiome model representing CF metacommunity diversity and benchmark on clinical data. We show that in the absence of antibiotics, replicate microbiome structures in a synthetic sputum medium are highly repeatable and dominated by oral commensals. In contrast, challenge with physiologically relevant antibiotic doses leads to substantial community perturbation characterized by multiple alternate pathogen-dominant states and enrichment of drug-resistant species. These results provide evidence that antibiotics can drive the expansion (via competitive release) of previously rare opportunistic pathogens and offer a path toward microbiome-informed conditional treatment strategies. IMPORTANCE We develop and clinically benchmark an experimental model of the cystic fibrosis (CF) lung infection microbiome to investigate the impacts of antibiotic exposures on chronic, polymicrobial infections. We show that a single experimental model defined by metacommunity data can partially recapitulate the diversity of individual microbiome states observed across a population of people with CF. In the absence of antibiotics, we see highly repeatable community structures, dominated by oral microbes. Under clinically relevant antibiotic exposures, we see diverse and frequently pathogen-dominated communities, and a nonevolutionary enrichment of antimicrobial resistance on the community scale, mediated by competitive release. The results highlight the potential importance of nonevolutionary (community-ecological) processes in driving the growing global crisis of increasing antibiotic resistance.
Keywords: antibiotic resistance; chronic infection; cystic fibrosis; experimental microbiome; infection microbiome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Lung function and microbiota diversity in cystic fibrosis.Microbiome. 2020 Apr 2;8(1):45. doi: 10.1186/s40168-020-00810-3. Microbiome. 2020. PMID: 32238195 Free PMC article.
-
Disruption of Cross-Feeding Inhibits Pathogen Growth in the Sputa of Patients with Cystic Fibrosis.mSphere. 2020 Apr 29;5(2):e00343-20. doi: 10.1128/mSphere.00343-20. mSphere. 2020. PMID: 32350096 Free PMC article.
-
High-Resolution Longitudinal Dynamics of the Cystic Fibrosis Sputum Microbiome and Metabolome through Antibiotic Therapy.mSystems. 2020 Jun 23;5(3):e00292-20. doi: 10.1128/mSystems.00292-20. mSystems. 2020. PMID: 32576651 Free PMC article.
-
Influence of the lung microbiome on antibiotic susceptibility of cystic fibrosis pathogens.Eur Respir Rev. 2019 Jul 8;28(152):190041. doi: 10.1183/16000617.0041-2019. Print 2019 Jun 30. Eur Respir Rev. 2019. PMID: 31285289 Free PMC article. Review.
-
Defining the Benefits of Antibiotic Resistance in Commensals and the Scope for Resistance Optimization.mBio. 2023 Feb 28;14(1):e0134922. doi: 10.1128/mbio.01349-22. Epub 2022 Dec 7. mBio. 2023. PMID: 36475750 Free PMC article. Review.
Cited by
-
Host tracheal and intestinal microbiomes inhibit Coccidioides growth in vitro.Microbiol Spectr. 2024 Jul 2;12(7):e0297823. doi: 10.1128/spectrum.02978-23. Epub 2024 Jun 4. Microbiol Spectr. 2024. PMID: 38832766 Free PMC article.
-
The impact of phage and phage resistance on microbial community dynamics.PLoS Biol. 2024 Apr 22;22(4):e3002346. doi: 10.1371/journal.pbio.3002346. eCollection 2024 Apr. PLoS Biol. 2024. PMID: 38648198 Free PMC article.
-
Host tracheal and intestinal microbiomes inhibit Coccidioides growth in vitro.bioRxiv [Preprint]. 2023 Oct 23:2023.10.23.563655. doi: 10.1101/2023.10.23.563655. bioRxiv. 2023. Update in: Microbiol Spectr. 2024 Jul 2;12(7):e0297823. doi: 10.1128/spectrum.02978-23. PMID: 37961490 Free PMC article. Updated. Preprint.
-
Gut microbiota interspecies interactions shape the response of Clostridioides difficile to clinically relevant antibiotics.PLoS Biol. 2023 May 11;21(5):e3002100. doi: 10.1371/journal.pbio.3002100. eCollection 2023 May. PLoS Biol. 2023. PMID: 37167201 Free PMC article.
-
Inference of dynamic interaction networks: A comparison between Lotka-Volterra and multivariate autoregressive models.Front Bioinform. 2022 Dec 22;2:1021838. doi: 10.3389/fbinf.2022.1021838. eCollection 2022. Front Bioinform. 2022. PMID: 36619477 Free PMC article.
References
-
- Laxminarayan R, Duse A, Wattal C, Zaidi AKM, Wertheim HFL, Sumpradit N, Vlieghe E, Hara GL, Gould IM, Goossens H, Greko C, So AD, Bigdeli M, Tomson G, Woodhouse W, Ombaka E, Peralta AQ, Qamar FN, Mir F, Kariuki S, Bhutta ZA, Coates A, Bergstrom R, Wright GD, Brown ED, Cars O. 2013. Antibiotic resistance: the need for global solutions. Lancet Infect Dis 13:1057–1098. doi:10.1016/S1473-3099(13)70318-9. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
