SCORe: SARS-CoV-2 Omicron Variant RBD-Binding DNA Aptamer for Multiplexed Rapid Detection and Pseudovirus Neutralization
- PMID: 35972202
- PMCID: PMC9397568
- DOI: 10.1021/acs.analchem.2c01993
SCORe: SARS-CoV-2 Omicron Variant RBD-Binding DNA Aptamer for Multiplexed Rapid Detection and Pseudovirus Neutralization
Abstract
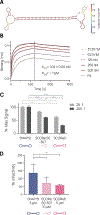
During the COVID-19 (coronavirus disease 2019) pandemic, several SARS-CoV-2 variants of concern emerged, including the Omicron variant, which has enhanced infectivity and immune invasion. Many antibodies and aptamers that bind the spike (S) of previous strains of SARS-CoV-2 either do not bind or bind with low affinity to Omicron S. In this study, we report a high-affinity SARS-CoV-2 Omicron RBD-binding aptamer (SCORe) that binds Omicron BA.1 and BA.2 RBD with nanomolar KD1. We employ aptamers SCORe.50 and SNAP4.74 in a multiplexed lateral flow assay (LFA) to distinguish between Omicron and wild-type S at concentrations as low as 100 pM. Finally, we show that SCORe.50 and its dimerized form SCOReD can neutralize Omicron S-pseudotyped virus infection of ACE2-overexpressing cells by >70%. SCORe therefore has potential applications in COVID-19 rapid diagnostics as well as in viral neutralization.
Figures



References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

