Developing Bottom-Up Induced Pluripotent Stem Cell Derived Solid Tumor Models Using Precision Genome Editing Technologies
- PMID: 35972367
- PMCID: PMC9529369
- DOI: 10.1089/crispr.2022.0032
Developing Bottom-Up Induced Pluripotent Stem Cell Derived Solid Tumor Models Using Precision Genome Editing Technologies
Abstract
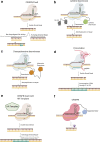
Advances in genome and tissue engineering have spurred significant progress and opportunity for innovation in cancer modeling. Human induced pluripotent stem cells (iPSCs) are an established and powerful tool to study cellular processes in the context of disease-specific genetic backgrounds; however, their application to cancer has been limited by the resistance of many transformed cells to undergo successful reprogramming. Here, we review the status of human iPSC modeling of solid tumors in the context of genetic engineering, including how base and prime editing can be incorporated into "bottom-up" cancer modeling, a term we coined for iPSC-based cancer models using genetic engineering to induce transformation. This approach circumvents the need to reprogram cancer cells while allowing for dissection of the genetic mechanisms underlying transformation, progression, and metastasis with a high degree of precision and control. We also discuss the strengths and limitations of respective engineering approaches and outline experimental considerations for establishing future models.
Conflict of interest statement
D.A.L. is the co-founder and co-owner of several biotechnology companies, including NeoClone Biotechnologies, Inc., Discovery Genomics, Inc. (recently acquired by Immusoft, Inc.), B-MoGen Biotechnologies, Inc. (recently acquired by Biotechne Corporation), and Luminary Therapeutics, Inc. D.A.L. holds equity in, serves as a Senior Scientific Advisor for and a Board of Director member for Recombinetics, a genome editing company. D.A.L. consults for Genentech, Inc., which is funding some of his research. W.A.W. is a co-founder of StemSynergy Therapeutics and holds equity in Nalo Therapeutics and Auron Therapeutics. The business of all these companies is unrelated to the contents of this article.
Figures


References
Publication types
MeSH terms
Grants and funding
- UH3 CA244687/CA/NCI NIH HHS/United States
- U54 CA243125/CA/NCI NIH HHS/United States
- P50 CA097257/CA/NCI NIH HHS/United States
- P30 CA082103/CA/NCI NIH HHS/United States
- R01 CA255369/CA/NCI NIH HHS/United States
- R01 AI146009/AI/NIAID NIH HHS/United States
- P01 CA254849/CA/NCI NIH HHS/United States
- U01 CA217864/CA/NCI NIH HHS/United States
- R01 AI161017/AI/NIAID NIH HHS/United States
- P01 CA217959/CA/NCI NIH HHS/United States
- R01 CA221969/CA/NCI NIH HHS/United States
- P50 CA136393/CA/NCI NIH HHS/United States
- R00 CA197484/CA/NCI NIH HHS/United States
- R01 NS106155/NS/NINDS NIH HHS/United States
- R01 NS115438/NS/NINDS NIH HHS/United States
- R21 CA237789/CA/NCI NIH HHS/United States
- R21 AI163731/AI/NIAID NIH HHS/United States

