Dynamic Catalysis Guided by Nucleic Acid Networks and DNA Nanostructures
- PMID: 35973134
- PMCID: PMC9853509
- DOI: 10.1021/acs.bioconjchem.2c00233
Dynamic Catalysis Guided by Nucleic Acid Networks and DNA Nanostructures
Abstract
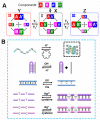
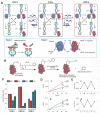
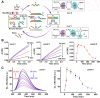
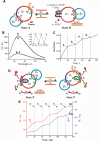
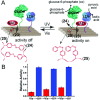
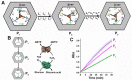
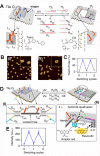
Nucleic acid networks conjugated to native enzymes and supramolecular DNA nanostructures modified with enzymes or DNAzymes act as functional reaction modules for guiding dynamic catalytic transformations. These systems are exemplified with the assembly of constitutional dynamic networks (CDNs) composed of nucleic acid-functionalized enzymes, as constituents, undergoing triggered structural reconfiguration, leading to dynamically switched biocatalytic cascades. By coupling two nucleic acid/enzyme networks, the intercommunicated feedback-driven dynamic biocatalytic operation of the system is demonstrated. In addition, the tailoring of a nucleic acid/enzyme reaction network driving a dissipative, transient, biocatalytic cascade is introduced as a model system for out-of-equilibrium dynamically modulated biocatalytic transformation in nature. Also, supramolecular nucleic acid machines or DNA nanostructures, modified with DNAzyme or enzyme constituents, act as functional reaction modules driving temporal dynamic catalysis. The design of dynamic supramolecular machines is exemplified with the introduction of an interlocked two-ring catenane device that is dynamically reversibly switched between two states operating two different DNAzymes, and with the tailoring of a DNA-tweezers device functionalized with enzyme/DNAzyme constituents that guides the dynamic ON/OFF operation of a biocatalytic cascade by opening and closing the molecular device. In addition, DNA origami nanostructures provide functional scaffolds for the programmed positioning of enzymes or DNAzyme for the switchable operation of catalytic transformations. This is introduced by the tailored functionalization of the edges of origami tiles with nucleic acids guiding the switchable formation of DNAzyme catalysts through the dimerization/separation of the tiles. In addition, the programmed deposition of two-enzyme/cofactor constituents on the origami raft allowed the dynamic photochemical activation of the cofactor-mediated biocatalytic cascade on the spatially biocatalytic assembly on the scaffold. Furthermore, photoinduced "mechanical" switchable and reversible unlocking and closing of nanoholes in the origami frameworks allow the "ON" and "OFF" operation of DNAzyme units in the nanoholes, confined environments. The future challenges and potential applications of dynamic nucleic acid/enzyme and DNAzyme conjugates are discussed in the conclusion paragraph.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





Similar articles
-
Active generation of nanoholes in DNA origami scaffolds for programmed catalysis in nanocavities.Nat Commun. 2019 Oct 31;10(1):4963. doi: 10.1038/s41467-019-12933-9. Nat Commun. 2019. PMID: 31672967 Free PMC article.
-
Switchable enzyme/DNAzyme cascades by the reconfiguration of DNA nanostructures.Chemistry. 2014 Dec 1;20(49):16203-9. doi: 10.1002/chem.201404122. Epub 2014 Oct 13. Chemistry. 2014. PMID: 25308317
-
Controlling the Catalytic Functions of DNAzymes within Constitutional Dynamic Networks of DNA Nanostructures.J Am Chem Soc. 2017 Jul 19;139(28):9662-9671. doi: 10.1021/jacs.7b04531. Epub 2017 Jul 10. J Am Chem Soc. 2017. PMID: 28627887
-
Dynamic Reconfigurable DNA Nanostructures, Networks and Materials.Angew Chem Int Ed Engl. 2023 Apr 24;62(18):e202215332. doi: 10.1002/anie.202215332. Epub 2023 Feb 14. Angew Chem Int Ed Engl. 2023. PMID: 36651472 Review.
-
Switchable and dynamic G-quadruplexes and their applications.Chem Soc Rev. 2022 Aug 30;51(17):7631-7661. doi: 10.1039/d2cs00317a. Chem Soc Rev. 2022. PMID: 35975685 Review.
Cited by
-
Adenosine-Triggered Dynamic and Transient Aptamer-Based Networks Integrated in Liposome Protocell Assemblies.J Am Chem Soc. 2025 Jun 4;147(22):19282-19295. doi: 10.1021/jacs.5c05090. Epub 2025 May 22. J Am Chem Soc. 2025. PMID: 40403280 Free PMC article.
-
Constitutional adaptation to pKa modulation by remote ester hydrolysis.Chem Sci. 2024 Apr 11;15(19):7092-7103. doi: 10.1039/d4sc01288g. eCollection 2024 May 15. Chem Sci. 2024. PMID: 38756812 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

