Antigenic cartography using sera from sequence-confirmed SARS-CoV-2 variants of concern infections reveals antigenic divergence of Omicron
- PMID: 35973428
- PMCID: PMC9353602
- DOI: 10.1016/j.immuni.2022.07.018
Antigenic cartography using sera from sequence-confirmed SARS-CoV-2 variants of concern infections reveals antigenic divergence of Omicron
Abstract
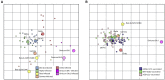
Large-scale vaccination campaigns have prevented countless hospitalizations and deaths due to COVID-19. However, the emergence of SARS-CoV-2 variants that escape from immunity challenges the effectiveness of current vaccines. Given this continuing evolution, an important question is when and how to update SARS-CoV-2 vaccines to antigenically match circulating variants, similarly to seasonal influenza viruses where antigenic drift necessitates periodic vaccine updates. Here, we studied SARS-CoV-2 antigenic drift by assessing neutralizing activity against variants of concern (VOCs) in a set of sera from patients infected with viral sequence-confirmed VOCs. Infections with D614G or Alpha strains induced the broadest immunity, whereas individuals infected with other VOCs had more strain-specific responses. Omicron BA.1 and BA.2 were substantially resistant to neutralization by sera elicited by all other variants. Antigenic cartography revealed that Omicron BA.1 and BA.2 were antigenically most distinct from D614G, associated with immune escape, and possibly will require vaccine updates to ensure vaccine effectiveness.
Keywords: Omicron; SARS-CoV-2; VOCs; antibodies; antigenic cartography; convalescent; neutralization; vaccination; variants of concern.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Barr I.G., Russell C., Besselaar T.G., Cox N.J., Daniels R.S., Donis R., Engelhardt O.G., Grohmann G., Itamura S., Kelso A., et al. WHO recommendations for the viruses used in the 2013–2014 Northern Hemisphere influenza vaccine: epidemiology, antigenic and genetic characteristics of influenza A(H1N1)pdm09, A(H3N2) and B influenza viruses collected from October 2012 to January 2013. Vaccine. 2014;32:4713–4725. doi: 10.1016/j.vaccine.2014.02.014. - DOI - PubMed
-
- Caniels T.G., Bontjer I., van der Straten K., Poniman M., Burger J.A., Appelman B., Lavell A.H.A., Oomen M., Godeke G.J., Valle C., et al. Emerging SARS-CoV-2 variants of concern evade humoral immune responses from infection and vaccination. Sci. Adv. 2021;7:eabj5365. doi: 10.1126/sciadv.abj5365. - DOI - PMC - PubMed
-
- Cele S., Karim F., Lustig G., San J.E., Hermanus T., Tegally H., Snyman J., Moyo-Gwete T., Wilkinson E., Bernstein M., et al. SARS-CoV-2 prolonged infection during advanced HIV disease evolves extensive immune escape. Cell Host Microbe. 2022;30:154–162.e5. doi: 10.1016/j.chom.2022.01.005. - DOI - PMC - PubMed
-
- Center for Disease Control and Prevention Antigenic characterization. 2021. https://www.cdc.gov/flu/about/professionals/antigenic.htm
-
- CoVariants Overview of variants in countries. 2022. https://covariants.org/per-country
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

