Identification of secreted proteins by comparison of protein abundance in conditioned media and cell lysates
- PMID: 35973625
- PMCID: PMC9756135
- DOI: 10.1016/j.ab.2022.114846
Identification of secreted proteins by comparison of protein abundance in conditioned media and cell lysates
Abstract
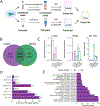
Analysis of the full spectrum of secreted proteins in cell culture is complicated by leakage of intracellular proteins from damaged cells. To address this issue, we compared the abundance of individual proteins between the cell lysate and the conditioned medium, reasoning that secreted proteins should be relatively more abundant in the conditioned medium. Marked enrichment for signal-peptide-bearing proteins with increasing conditioned media to cell lysate ratio, as well loss of this signal following brefeldin A treatment, confirmed the sensitivity and specificity of this approach. The subset of proteins demonstrating increased conditioned media to cell lysate ratio in the presence of Brefeldin A identified candidates for unconventional secretion via a pathway independent of ER to Golgi trafficking.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F, Proteomics. Tissue-based map of the human proteome, Science 347 (2015), 1260419. - PubMed
-
- Uhlen M, Karlsson MJ, Hober A, Svensson AS, Scheffel J, Kotol D, Zhong W, Tebani A, Strandberg L, Edfors F, Sjostedt E, Mulder J, Mardinoglu A, Berling A, Ekblad S, Dannemeyer M, Kanje S, Rockberg J, Lundqvist M, Malm M, Volk AL, Nilsson P, Manberg A, Dodig-Crnkovic T, Pin E, Zwahlen M, Oksvold P, von Feilitzen K, Haussler RS, Hong MG, Lindskog C, Ponten F, Katona B, Vuu J, Lindstrom E, Nielsen J, Robinson J, Ayoglu B, Mahdessian D, Sullivan D, Thul P, Danielsson F, Stadler C, Lundberg E, Bergstrom G, Gummesson A, Voldborg BG, Tegel H, Hober S, Forsstrom B, Schwenk JM, Fagerberg L, Sivertsson A, The human secretome, Sci. Signal 12 (2019). - PubMed
-
- Hebert DN, Molinari M, In and out of the ER: protein folding, quality control, degradation, and related human diseases, Physiol. Rev 87 (2007) 1377–1408. - PubMed
-
- Zanetti G, Pahuja KB, Studer S, Shim S, Schekman R, COPII and the regulation of protein sorting in mammals, Nat. Cell Biol 14 (2011) 20–28. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

