Modeling PRPF31 retinitis pigmentosa using retinal pigment epithelium and organoids combined with gene augmentation rescue
- PMID: 35974011
- PMCID: PMC9381579
- DOI: 10.1038/s41536-022-00235-6
Modeling PRPF31 retinitis pigmentosa using retinal pigment epithelium and organoids combined with gene augmentation rescue
Abstract
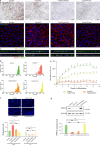
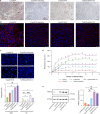
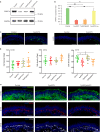
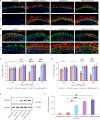
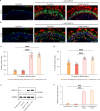
Mutations in the ubiquitously expressed pre-mRNA processing factor (PRPF) 31 gene, one of the most common causes of dominant form of Retinitis Pigmentosa (RP), lead to a retina-specific phenotype. It is uncertain which retinal cell types are affected and animal models do not clearly present the RP phenotype observed in PRPF31 patients. Retinal organoids and retinal pigment epithelial (RPE) cells derived from human-induced pluripotent stem cells (iPSCs) provide potential opportunities for studying human PRPF31-related RP. We demonstrate here that RPE cells carrying PRPF31 mutations present important morphological and functional changes and that PRPF31-mutated retinal organoids recapitulate the human RP phenotype, with a rod photoreceptor cell death followed by a loss of cones. The low level of PRPF31 expression may explain the defective phenotypes of PRPF31-mutated RPE and photoreceptor cells, which were not observed in cells derived from asymptomatic patients or after correction of the pathogenic mutation by CRISPR/Cas9. Transcriptome profiles revealed differentially expressed and mis-spliced genes belonging to pathways in line with the observed defective phenotypes. The rescue of RPE and photoreceptor defective phenotypes by PRPF31 gene augmentation provide the proof of concept for future therapeutic strategies.
© 2022. The Author(s).
Conflict of interest statement
A.S.-B., S.R., J.-A.S., and O.G. are inventors on patents on hiPSC retinal differentiation and on the use of hiPSC retinal derivatives to treat retinal degeneration, licensed to Gamut Cell Tx. L.C.B. is an inventor of patent applications for AAV capsid variants and AAV screening methods, and she is a founder of Avista Therapeutics and Vegavect.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

