A context-dependent and disordered ubiquitin-binding motif
- PMID: 35974206
- PMCID: PMC9381478
- DOI: 10.1007/s00018-022-04486-w
A context-dependent and disordered ubiquitin-binding motif
Abstract
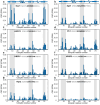
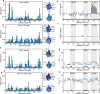
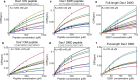
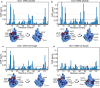
Ubiquitin is a small, globular protein that is conjugated to other proteins as a posttranslational event. A palette of small, folded domains recognizes and binds ubiquitin to translate and effectuate this posttranslational signal. Recent computational studies have suggested that protein regions can recognize ubiquitin via a process of folding upon binding. Using peptide binding arrays, bioinformatics, and NMR spectroscopy, we have uncovered a disordered ubiquitin-binding motif that likely remains disordered when bound and thus expands the palette of ubiquitin-binding proteins. We term this motif Disordered Ubiquitin-Binding Motif (DisUBM) and find it to be present in many proteins with known or predicted functions in degradation and transcription. We decompose the determinants of the motif showing it to rely on features of aromatic and negatively charged residues, and less so on distinct sequence positions in line with its disordered nature. We show that the affinity of the motif is low and moldable by the surrounding disordered chain, allowing for an enhanced interaction surface with ubiquitin, whereby the affinity increases ~ tenfold. Further affinity optimization using peptide arrays pushed the affinity into the low micromolar range, but compromised context dependence. Finally, we find that DisUBMs can emerge from unbiased screening of randomized peptide libraries, featuring in de novo cyclic peptides selected to bind ubiquitin chains. We suggest that naturally occurring DisUBMs can recognize ubiquitin as a posttranslational signal to act as affinity enhancers in IDPs that bind to folded and ubiquitylated binding partners.
Keywords: Context; Cyclic peptide; Deep mutational scanning; IDP; NMR; SLiM; UBM; Ubiquitin.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no conflicts of interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

